Protocol modifications capture viral RNA in single cell and spatial samples
The Innovator Blog Series celebrates research conducted by 10x Genomics customers who have demonstrated scientific ingenuity by adapting Chromium Single Cell or Visium Spatial sequencing-based assays. These innovations are distinguished by their originality and potential impact on scientific discovery, including providing access to novel analytes, advancing multiomic analysis techniques, and demonstrating critical applications to human health and disease research. These techniques are customer developed, meaning they are not officially supported by 10x Genomics.
The research in this article was featured in our recent Immunology Spotlight Webinar. Learn more here.
*Editor's Note: The method described in this article, spatial total RNA-sequencing (STRS), has now been published in Nature Biotechnology.*
Why use multiomics for infectious disease research?
Viral pathogenesis represents a unique intersection of single cell and spatial biology. At different points in the process of infection and with varying susceptibility, host cells are invaded by viral agents, resulting in dysregulated transcriptomic signatures, functional consequences, and even cell death. Simultaneously, viral dissemination is aided and accomplished through the close interaction of infected and non-infected host cells in the tissue microenvironment.
Some viruses have evolved to stabilize existing cellular interactions and even create new ones to promote transmission, such as the Herpes simplex virus which recruits nerve endings and drives skin cell migration to enable cell-contact formation (1). In these ways, the presence of virus could fundamentally alter cellular colocalization in tissue.
But the influence flows in two directions. Viral dissemination is itself highly influenced by the organization of the host tissue systems:
“Local and systemic virus transmission is critically influenced by the tissue physiology. The mechanism of virus transmission is shaped by the tissue context and influenced by physical barriers such as the fluid-tissue interfaces (lymph/lymph node, blood/spleen), local cell populations with limited exchange with the systemic cell pool or spatially restricted cell migration and cell-cell interaction” (1).
Understanding viral pathogenesis and advancing antiviral strategies therefore require a holistic view of both the single cell and spatial dynamics of infection. This is the basis for a multiomic approach to investigate infectious disease. This approach allows scientists to view the underlying biology of an infected tissue from multiple perspectives, including capturing multiple analytes—such as RNA, protein, or clonotype information—simultaneously from single cells or a tissue sample, or integrating different analytes from multiple technologies.
“Multiomics can provide researchers with a greater understanding of the flow of information, from the original cause of disease (genetic, environmental, or developmental) to the functional consequences or relevant interactions” (2).
Integrating single cell and spatial to advance myocarditis insights
A tragic loss may break our hearts, but the human heart can suffer more than emotional injuries. Viral-induced myocarditis is a condition in which the heart muscles, or myocardium, become inflamed as a result of viral infection and influx of host immune cells into the muscle. This leads to tissue scarring and organ dysfunction, which can disrupt the pumping rhythm of the heart and reduce blood flow (3). Though classified as a rare condition, there were 3.1 million diagnosed cases of myocarditis in 2017 (4). The condition is often seen in otherwise healthy, young people, representing the third-leading cause of sudden death in children and young adults (4). Moreover, 2020–2021 COVID-19 patient data suggested an association between SARS-CoV-2 infection and myocarditis, pointing to heart inflammation and scarring as another COVID-19–related complication (5).
In his presentation at our recent Immunology Spotlight Webinar, Madhav Mantri, PhD candidate from the Meinig School of Biomedical Engineering, Cornell University, described the dangers of viral-induced myocarditis. Using a mouse model of Reovirus Type-1-lang infection, which enters mice through the intestine and moves on to infect multiple organs, including the heart, Mantri and colleagues sought to explore viral pathogenesis at the cellular and tissue level:
- What are the steps that drive broader viral transmission, as well as increasing inflammatory severity in the heart?
- What is the cellular and spatial composition of myocarditic tissues?
This inquiry led them to use Chromium Single Cell Gene Expression to first atlas the cells present in infected ileum and heart tissues from Day 1 to Day 10 post-infection. Here, a modification to the single cell RNA-seq protocol allowed Mantri and colleagues to define the specific cell types targeted by reovirus in both tissues. Their modification, called xGen reovirus transcript enrichment, applied a panel of biotinylated probes to the single cell RNA-seq libraries to pull down viral transcripts, which were then amplified, resequenced, and quantified.
A natural logic of viral transmission emerged: on Day 1, enteroendocrine cells, which act as a barrier between the intestinal environment and gut tissue, were the most infected. By Day 4, viral transcripts appeared primarily in lymphatic endothelial cells inside the gut villi, from which virus could then leak into the bloodstream. By Day 7, heart endothelial cells (which line the heart vasculature), and endocardial cells (which compose the walls of the heart) were most infected, confirming the cells most vulnerable to infection were those at the fluid–tissue interfaces.
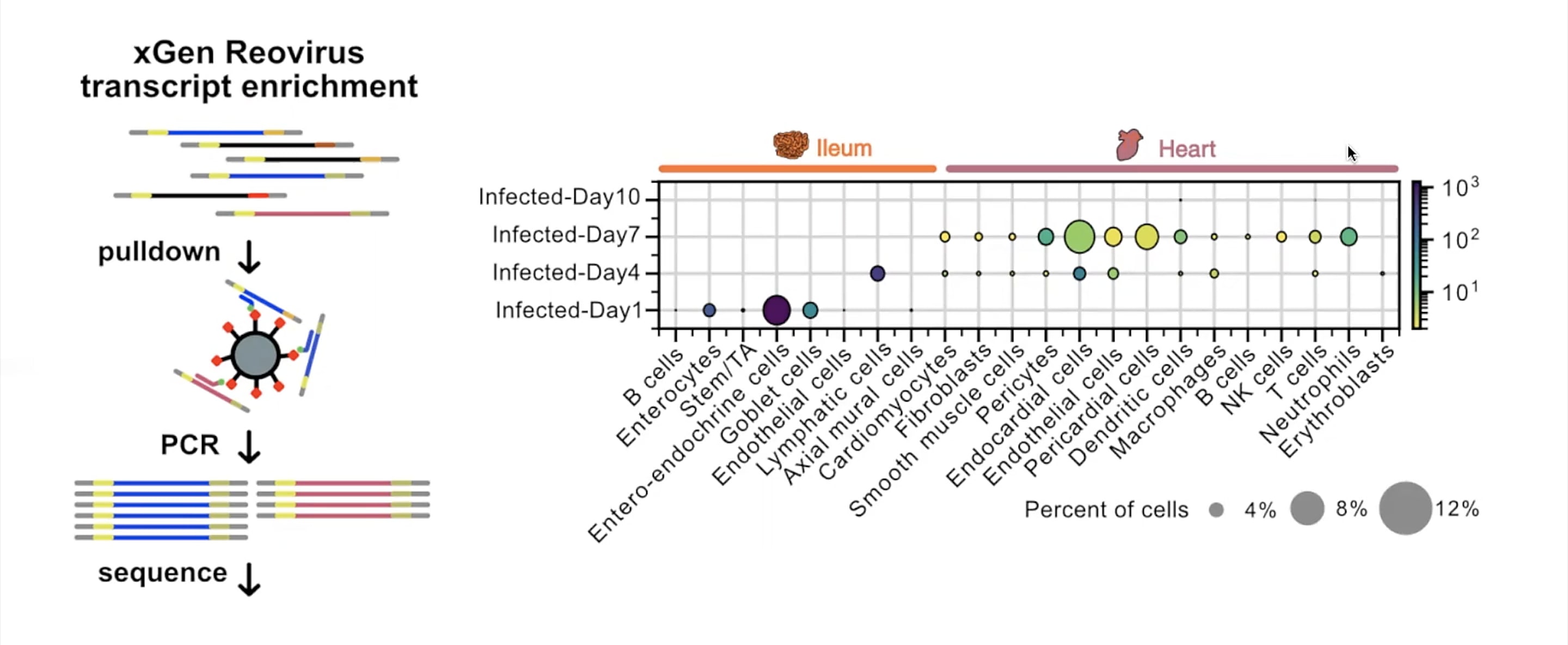
To determine how infected cells coordinated with the host immune response to drive inflammatory severity in the heart, Mantri and his team focused on endothelial populations, whose transcriptomic profiles suggested a robust immune response. At Day 7 post-infection, endothelial cells expressed chemokines CXCL9 and CXCL10, which are involved in T-cell recruitment, and CD74, an MHC class protein involved in antigen presentation. Endothelial cells also had upregulated expression of caspase genes, indicators of cell death, suggesting infected endothelial cells were simultaneously presenting antigen to infiltrating T cells and the target of T-cell induced death.
To round out these findings, Mantri and colleagues investigated whether or not endothelial and immune cells colocalized in the morphology of myocarditic scar tissue. This led them to analyze heart sections with Visium Spatial Gene Expression, then integrate the spatial data with single cell data to deconvolute and calculate specific cell-type proportions within individual spots of the spatial transcriptomics data. They found that CXCL9-expressing endothelial cells, cytotoxic T cells, and dendritic cells were enriched and colocalized in myocarditic patches and adjacent border zones. Based on this evidence of a strong presence of cytotoxic T cells in the scar tissue, they concluded that the host immune response was itself the greater driver of tissue damage and must exist in a delicate balance between under- and overcompensating in response to viral infection.
Read more about this study here →
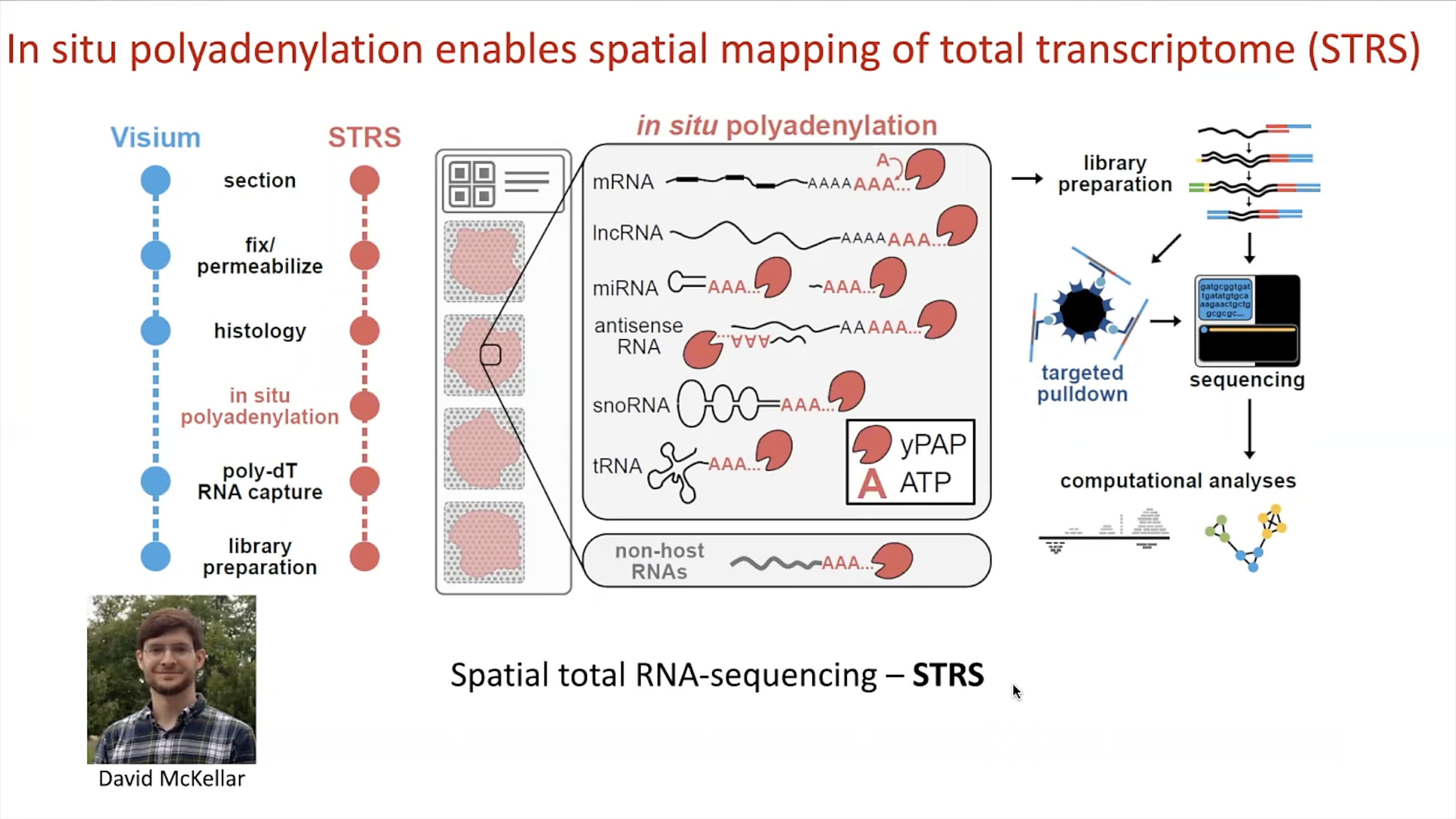
In situ polyadenylation powers spatial total RNA-sequencing
The team’s innovations didn’t stop at pulling viral RNA transcripts from single cell gene expression data. In line with their multiomic, multitechnology approach to the study of myocarditis, Mantri, in collaboration with PhD candidate David McKellar, developed a method called spatial total RNA-sequencing (STRS). A modification of the Visium Spatial assay, STRS allows spatial mapping of total transcriptomes in tissue, including endogenous polyadenylated host mRNA, regulatory non-coding RNA, and non-polyadenylated viral RNA.
While the standard Fresh Frozen Visium assay would see a dropout of non-polyadenylated reovirus RNA, the team modified the protocol to include an in situ polyadenylation step on the sectioned tissue, allowing them to add a polyA tail onto viral RNA transcripts. Those transcripts, in addition to a number of other RNA molecules that lack the polyA tail, could then hybridize with poly-dT RNA capture sequences on the Visium slides in subsequent steps of the workflow. Incorporating a similar approach as that of their xGen reovirus transcript enrichment method for single cell libraries, a targeted pulldown of these RNA transcripts from spatial sequencing libraries could then ensure quantification of viral RNA while maintaining spatial information.
Using STRS, Mantri and McKellar were able to show that foci of reovirus RNA in infected heart tissue sections overlapped with inflammation patches clearly identified morphologically by H&E staining.
Learn more about spatial total RNA-sequencing →
Pushing the boundaries of multiomics
These innovations represent an important advancement for infectious disease research, providing access to viral RNA in both single cell and spatial sequencing applications and expanding the capabilities of already powerful tools. Moreover, they open the door to deeper insights into viral pathogenesis at the cellular and tissue level, promising great advances in antiviral strategies that can alleviate life-threatening conditions such as myocarditis.
We are proud to recognize these methods as part of our Innovator Series, and we continue to be amazed at the ways scientists in our community are driving the leading edge of multiomic techniques. To learn more about the research featured in this article, and to hear a live Q&A with our speaker Madhav Mantri, watch the on-demand Immunology Spotlight Webinar.
References:
- Sewald X, et al. Viruses exploit the tissue physiology of the host to spread in vivo. Curr Opin Cell Biol 41: 81–90 (2016). doi: 10.1016/j.ceb.2016.04.008
- Hasin Y, et al. Multi-omics approaches to disease. Genome Biol 18: 83 (2017). doi: 10.1186/s13059-017-1215-1
- https://www.hopkinsmedicine.org/health/conditions-and-diseases/myocarditis
- https://www.myocarditisfoundation.org/about-myocarditis/
- https://www.cdc.gov/mmwr/volumes/70/wr/mm7035e5.htm
