Not a losing battle: Single cell tools provide new insights into chronic autoimmune diseases
March is National Autoimmune Disease Awareness Month, and for the millions of people who suffer from a disease with no cure, education is key to advances in research and therapies. In this blog post, we highlight two publications that use single cell tools to study the origins of multiple sclerosis and Crohn’s disease. Both studies were launched with the aim of learning more about how these diseases develop by comparing single cell states in disease to healthy immune system surveillance and normal embryonic development, respectively. Our hope is to promote not only awareness of autoimmunity-related diseases but also showcase the higher potential of applying 10x Genomics single cell and spatial solutions to impact human health.
An autoimmune disease develops when a person’s immune system mistakes the body’s own healthy cells for foreign cells and attacks them. Autoimmune diseases can affect the body’s organs, tissues, and cells. There are more than 80 autoimmune diseases, the most common including type 1 diabetes, rheumatoid arthritis, systemic lupus erythematosus, and inflammatory bowel disease (1). Others, less common but equally vexing, are multiple sclerosis (MS) and Crohn’s disease. According to the NIH, up to 23.5 million Americans live with an autoimmune disease (2).
Researchers don’t know exactly what causes autoimmune diseases, but they do know that your genes may make you more susceptible while environmental factors can trigger or exacerbate an underlying tendency. The process of diagnosing an autoimmune disease is often referred to as a “diagnostic odyssey” in that it can take several visits to many doctors, typically over the course of years, before a diagnosis is made. Increasing our understanding of the root cause of an autoimmune disease is critically important to improving detection and diagnosis, which will ultimately lead to improved treatment and patient outcomes.
Key to this understanding is shedding light on disease mechanisms—what genes could be targeted by therapies, or what proteins might be used as diagnostic or prognostic biomarkers—and this work starts with examining the biology of healthy individuals. For example, deciphering healthy immune system surveillance at the single cell, tissue-specific level is one way to uncover potential disease mechanisms. Tracking the process of normal embryonic development to uncover differentiation factors that may be linked to the root cause of an autoimmune-related disease is another promising tactic. In this post, we look at two studies that employed 10x Genomics single cell technologies to examine what underlies the biology of MS and Crohn’s disease.
Defining health: characterizing immune cells in the CSF for insight into MS
In MS, the immune system attacks the protective sheath, called myelin, covering nerve cell axons, resulting in damage or deterioration of the nerve cells. Symptoms of MS range from muscle spasms, stiffness, and weakness to losing the ability to walk independently or at all. There's no cure for multiple sclerosis, but treatments can help manage symptoms.
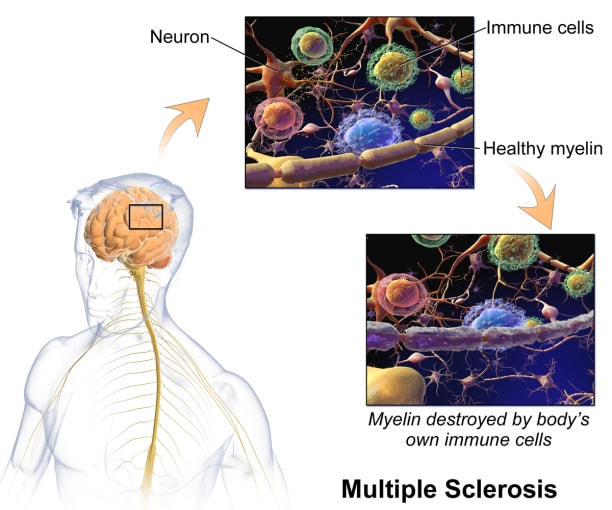
When it comes to the immune system and the brain, it was previously thought that the central nervous system (CNS) was immune privileged, meaning immune cells inhabiting the body can’t enter the CNS. Now, however, we know that there is active interaction between the peripheral (or, systemic) immune cells and the CNS—largely through the main point of entry, the cerebrospinal fluid (CSF). And while these cells play a crucial role in protecting the CNS from pathogens, peripheral immune cells that enter the CNS can also drive neuroinflammation. For example, in MS, there is evidence of immune activation in the CSF, which is used for diagnosis (3). Characterizing CSF T cells in healthy people not only offers tissue-specific context to T-cell profiles in disease states like MS, where there is neuroinflammation, but it also provides a way to look at immune function in the CNS in health and disease.
In a recent study (4) led by Jenna L. Pappalardo, PhD, in the lab of David Hafler, MD, at the Yale School of Medicine, she and her team used a combination of single cell RNA-sequencing (scRNA-seq) and paired T-cell receptor (TCR) sequencing to characterize T-cell state and activity in the healthy CSF compared to that of newly diagnosed (within 12 months) relapsing-remitting MS patients. Performing scRNA-seq on 16,752 T cells from the blood and 14,043 T cells from the CSF of six healthy individuals, her team uncovered four subsets in the blood—naïve CD4, naïve CD8, CD4 memory, and CD8 memory—and four in the CSF—three clusters of memory CD4 T cells and one memory CD8 cluster. Analyses showed that, compared to peripheral blood, the healthy CSF is enriched for CD4 T cells with Th1 function, including a CD4 subtype with cytotoxic capability. Compared to cells from CD8 clusters in the blood, cells in the CD8 memory cluster in the CSF had increased expression of cytotoxicity-related genes.
Using paired TCR sequencing, they then characterized clonally expanded T cells in healthy CSF and compared these profiles to clonally expanded T cells obtained from the CSF of five patients with MS (54,676 total blood and CSF cells). While they did not see an overall difference in clonally expanded conventional T cells in the CSF, they did find that CSF T cells in patients with MS have increased expression of genes related to T-cell activation and cytotoxicity.
In summary, characterizing T cells in the CNS may help elucidate the mechanisms behind how these cells perform immune surveillance without inflicting harmful inflammation in the brain and spinal cord. It could also point to roles for these immune cells in regulating autoimmune, neurodegenerative, anti-tumor, and anti-pathogen immune responses within the CNS.
Mapping development to dysfunction in Crohn’s disease
Crohn's disease is a type of inflammatory bowel disease (IBD) that leads to inflammation of the digestive tract. Symptoms include abdominal pain, diarrhea, fatigue, weight loss, and malnutrition. While there's no known cure for Crohn's disease, therapies can lessen symptoms and lead to remission and healing of inflammation.
Environmental factors during early development have been implicated in varied immune-related pathologies, including IBD. Improved understanding of human intestinal maturation will help make connections between healthy and disease states of the intestinal tract and be a critical step toward treatments. To that end, a study (5) led by teams from the labs of Sarah Teichmann, PhD, at the Wellcome Sanger Institute and Matthias Zilbauer, PhD, at Cambridge University looked to fetal development for clues to what goes awry in this disease.
To start, they generated a single cell map of the developing human intestine. They performed single cell transcriptional profiling of human embryonic and early fetal gut samples obtained from nine human embryos spanning 6 to 10 weeks post-conception. Additionally, they profiled mucosal biopsies from the small bowel of healthy children between 4 and 12 years old and a group of children newly diagnosed with Crohn’s disease. In total, they studied approximately 90,000 primary human intestinal cells.
This analysis revealed seven major cell types in embryonic/fetal samples; differences in cellular composition across the three developmental stages (embryonic, fetal, and pediatric); and differentiated cell states more frequently seen in the proximal than distal regions of the developing gut. Sub-clustering analysis showed that, at the embryonic stages, the epithelium is composed of high-cycling, uniform progenitors that express low levels of LGR5 (leucine-rich repeat-containing G-protein coupled receptor 5), a canonical adult stem cell gene.
When they compared scRNA-seq profiles from seven pediatric Crohn’s disease epithelial samples to matched healthy controls, they observed disease-associated changes in the epithelial composition. Interestingly, they discovered a reactivation of fetal transcription factors in the pediatric Crohn’s disease cells, including TP53, MYC, HMGA1, and HMGA2, to name a few.
In short, their work exemplifies the impact that visualizing fetal development at the single cell level can have on furthering our understanding of Crohn’s disease. Their gut cell atlas will be a useful tool for other researchers and is a welcome addition to the growing Human Cell Atlas.
Laying the groundwork for better treatments for autoimmune diseases
Insights gleaned from studying healthy tissue at the single cell level will go a long way toward laying the groundwork for translational studies and clinical trials for autoimmune diseases. As we understand more about how cell subtype and state impact the complex interaction of the peripheral immune system with the CNS in MS, and why diseases like Crohn’s resemble early developmental states when looking at single cell factors, scientists will be able to build off this basic knowledge to conduct more advanced studies aimed at deciphering what goes awry in autoimmune diseases and using these targets to develop new therapies.
Learn more about how 10x Genomics single cell and spatial technologies are supporting immunology research.
References
- https://www.niaid.nih.gov/diseases-conditions/autoimmune-diseases
- NIH Autoimmune Diseases Coordinating Committee: Autoimmune Diseases Research Plan, March 2005. https://www.niaid.nih.gov/sites/default/files/adccfinal.pdf (Last accessed October 10, 2017)
- Link H and Huang Y-M. Oligoclonal bands in multiple sclerosis cerebrospinal fluid: An update on methodology and clinical usefulness. J Neuroimmunol 180: 17–28, 2006.
- Pappalardo JL, et al. Transcriptomic and clonal characterization of T cells in the human central nervous system. Sci Immunol 5: eabb8786, 2020.
- Elmentaite R, et al. Single-cell sequencing of developing human gut reveals transcriptional links to childhood Crohn’s disease. Dev Cell 55: 771–783.e5, 2020.
