Paving new roads in the quest for novel Alzheimer’s Disease drug targets
The failure of many clinical trial drug candidates for Alzheimer’s Disease targeting amyloid-beta plaques is renewing efforts to study brain cells at a more fundamental level. Here we highlight recent publications that use single cell RNA-seq as a valuable tool in basic research for high resolution characterization of non-neuronal cells in AD and preclinical assessment of a therapeutic candidate.
Our scientific understanding of Alzheimer’s Disease (AD) has grown enormously since it was first described in 1906. However, there is still a lot of uncertainty in predicting how the disease will progress in each individual. For example, we don’t know why some individuals can live with the disease as long as 20 years following diagnosis, despite the average post-diagnosis survival rate being much lower at 4 to 8 years (1). Earlier in 2020, our gaps in understanding surfaced with the clinical trial failures of not one, but two therapeutics targeting the beta-amyloid (Aβ) protein, whose buildup is the principal clinical manifestation of AD (2). These failures, and those of many other previous candidates that have not lessened cognitive decline, are leading some researchers back to the bench to better understand the types of cells involved in AD development and identify new drug targets (3).
Tracing astrocyte gene expression changes in AD progression
Non-neuronal cells have been reported to play a role in Alzheimer’s Disease onset and progression. Still, the mechanism is not yet understood, partly because the heterogeneity of these cells can make traditional bulk analyses challenging (4, 5). Recognizing this fundamental gap, a coalition of researchers from The Hebrew University of Jerusalem, The Weizmann Institute, The Broad Institute, and MIT joined forces to build detailed cellular-molecular maps of the hippocampus to characterize the role non-neuronal cell populations from this brain region play in AD (5).
To build these maps, the researchers performed RNA-seq with Chromium Single Cell Gene Expression to profile 54,769 single nuclei derived from the hippocampi of wild-type mice or a transgenic AD mouse model. Their comparative analysis revealed that astrocytes could be categorized into three major transcriptional states, with one of those states being unique to the AD model. A time point analysis revealed that the unique AD-specific subpopulation, termed disease-associated astrocytes (DAA), appeared before manifestations of cognitive decline and increased in prevalence through the time points, reflecting more advanced disease states. Furthermore, this subpopulation was localized in proximity to Aβ plaques in regions of the brain where disease manifestation is most severe.
While animal models provide a great window into disease mechanisms, the findings derived from these models don’t always translate to the human condition. This study took the guesswork out of the human relevance of these results, finding equivalents of the three different transcriptional states in aging postmortem brains and a higher frequency of DAA-like cells in patients with AD. These findings are suggestive of a dynamic astrocyte activation process in Alzheimer’s, particularly in the initial pathogenesis, and hint that DAAs may be a source for novel therapeutic targets.
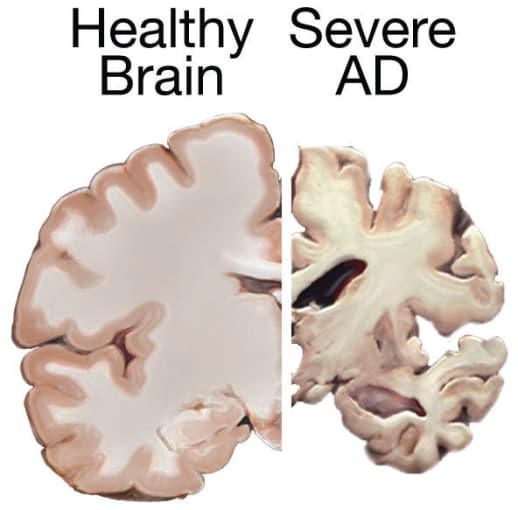
Therapeutic potential of TREM2: from bench findings to preclinical verification
Microglia are another population of non-neuronal cells that researchers have been investigating closely. Variants of a receptor protein, triggering receptor expressed on myeloid cells 2 (TREM2), are associated with increased risk of Alzheimer’s Disease (6). In a January 2020 Nature Medicine article, a cohort of international researchers led by senior investigator Marco Colonna (Washington University School of Medicine) reported a comprehensive gene expression study examining transcriptional changes in the disease associated with TREM2 in mouse and human brains (7).
Intriguingly, the single nuclei sequencing performed with Chromium Single Cell Gene Expression revealed that the transcriptional signatures expressed in AD pathologies of the mouse model and human brain were quite distinct. The mouse models of AD contained a unique population of activated microglia, termed disease-associated microglia (DAM), whose presence is Aβ- and TREM2-dependent. In contrast, human AD samples contained a microglia population with a transcriptional signature similar to those observed in peripheral nerve injury. But that is not where the TREM2 story ends. The study also uncovered that patients expressing the R47H and R62H variants of TREM2 had defective microglial activation, suggesting that TREM2 still makes an essential contribution in human AD pathogenesis.
These findings, along with other promising basic research studies, raised the intriguing possibility that TREM2 would make a good therapeutic target. To examine this possibility, members of the Colonna Laboratory teamed up with Alector LLC, a biopharmaceutical company focused on neurodegenerative diseases, for a preclinical examination of AL002c, an anti-human TREM2 agonistic mAb, as a modulator of microglial activation and, therefore, AD (8). After confirmation that AL002c is capable of activating hTREM2, Chromium Single Cell Gene Expression was used to identify how acute administration of this therapeutic impacted microglial activation.
Based on gene expression profiles, the microglial cells (n = 31,948) were subdivided into homeostatic microglia, stage 1 DAM, transitioning microglia, proliferating microglia, and microglia expressing IFN-induced genes. Acute administration of AL002c into mice reduced the number of microglia with homeostatic transcription profiles while increasing the number of transitioning and proliferating microglia, both of which are new subtypes that had not previously been identified. This induced proliferation was also seen in transgenic mice containing TREM2 variants.
Further experiments demonstrated generally positive outcomes in response to sustained AL002c treatment, including reduced filamentous Aβ plaques and neurite damage, reduction of impacted behavior, and a lessened microglial inflammatory response. These preclinical assessments and promising safety results from a first-in-human phase I clinical trial suggest that AL002c is a promising AD therapy candidate that should be investigated further.
Furthering our understanding of AD
Although scientists are still working to uncover the molecular underpinnings of Alzheimer’s Disease, these studies show that they are well-equipped to tackle the biological complexities of the disease with powerful tools like single cell RNA-seq. As the genetic and functional differences of disease-associated cell subpopulations are better characterized, scientists will be able to address questions that were not previously accessible, breaking down barriers that once hindered our complete understanding of AD. These basic discoveries can then be used to evaluate the preclinical feasibility of new, more promising drug targets.
References
- https://www.alz.org/alzheimers-dementia/facts-figures
- https://www.advisory.com/daily-briefing/2020/02/12/alzheimers-disease
- https://www.npr.org/sections/health-shots/2020/06/25/883026917/alzheimer-s-research-is-going-back-to-basics-to-find-best-way-forward
- BD Strooper, E Karran. The Cellular Phase of Alzheimer’s Disease. Cell. 164, 603–615 (2016).
- N Habib et al., Disease-Associated Astrocytes in Alzheimer’s Disease and Aging. Nat. Neurosci. 23, 701–706 (2020).
- T K Ulland, M Colonna. TREM2 — A Key Player in Microglial Biology and Alzheimer Disease. Nat. Rev. Neurol. 14, 667–675 (2018).
- Y Zhou et al., Human and Mouse Single-Nucleus Transcriptomics Reveal TREM2-Dependent and TREM2-Independent Cellular Responses in Alzheimer’s Disease. Nat. Med. 26, 131–142 (2020).
- S Wang et al., Anti-Human TREM2 Induces Microglia Proliferation and Reduces Pathology in an Alzheimer’s Disease Model. J. Exp. Med. 217, e20200785 (2020).
