Sample Preparation Tips for Your Single-cell RNA-seq Experiments
Single-cell RNA-sequencing (scRNA-seq) is a transformative technology that provides unprecedented insight into complex biological systems, allowing researchers to explore gene expression dynamics on a cell-by-cell basis. But to get the most out of this powerful tool, you’ll need to know how to properly prepare your samples.
Fortunately, we recently published a Bench Tips article with Biocompare that tells you all about how to best prepare samples for scRNA-seq.
Some of the highlights from the article include:
Optimize for your cell type.
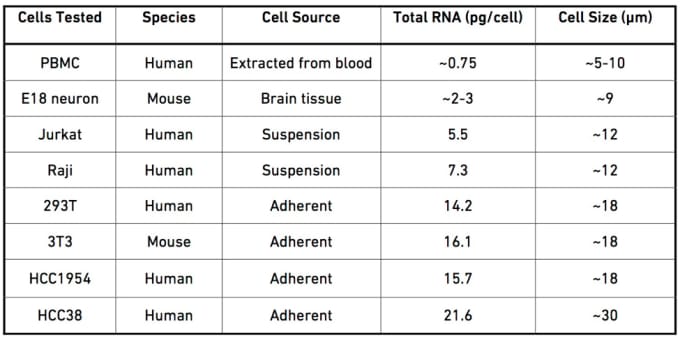
Every sample type has its own set of limitations and requirements, and knowing these in advance will allow you to optimize your sample prep and ultimately result in more robust data. For example, suspension cell lines, bead-enriched cells, and flow-sorted cells are already suspended and only need to be washed and counted before use with the Chromium™ Single Cell 3’ Solution.
Additionally, you should keep in mind that different cell types display a wide range of initial RNA content, and an accurate measure of the RNA input will impact key decisions throughout the workflow, such as the number of PCR cycles used in library preparation.
Pipette carefully.
Pipetting technique is critical for maintaining the quality of your single-cell sample. Immediately before loading your sample onto a microfluidic chip, it should be pipette-mixed gently with a wide-bore pipette tip. It is crucial that the cells are handled gently during this step as mixing roughly can cause premature cell lysis and create high ambient RNA levels that contribute to background noise in the system. Additionally, during transfer, your sample should be aspirated from the middle of the tube each time in order to pull consistent numbers of cells. As the sample settles, pulling from either the top or the bottom of the solution can cause you to aspirate very different numbers of cells.
Count your cells.
After washing and straining, use a cell counting device, such as a hemocytometer or a Countess® II FL Automated Cell Counter, to take an accurate count of the cells in your sample. This step is crucial as it will allow you calculate the volume of cell suspension needed to load the targeted number of cells.
Remove dead cells.
The presence of too many dead cells in your sample might affect your targeted cell recovery number so it’s important to reduce the percentage of dead cells in a single-cell suspension before sequencing. We recommend using our Demonstrated Protocol on Removal of Dead Cells from Single Cell Suspensions for best results.
Read our full Bench Tips article to learn more about preparing optimized single-cell samples. We also have a number of sample preparation protocols available on our support site for further guidance.
