Single Cell Gene Expression Flex: Lock in cell states, unlock potential
Since the writing of this blog, we have introduced GEM-X Flex Gene Expression which has lower input requirements and additional performance gains. Click here to learn more.
Over the past decade, single cell technology has given researchers the ability to probe further, dive deeper, and answer more complex questions about the biological systems that drive human health and disease. Since its debut in 2009 (1, 2), scientists have put single cell RNA sequencing to good use, discovering a wealth of information in immunology (3, 4), cancer (5, 6), neuroscience (7, 8), and many other research areas. The allure and potential of single cell sequencing lies in the ability to view biology with enough resolution to enable the differentiation of rare cell types, the discovery of new ones, and the deconvolution of hidden heterogeneity.
But what do you do when the answers to your questions lie in cells that are out of reach due to the timing or location of sample collection? What if you could overcome the logistical challenges in sample handling that stand between you and novel biological insights? At our recent Xperience 2022 event, we were excited to share more details about Single Cell Gene Expression Flex, a new assay that addresses these concerns, bringing sample fixation and expanded sample accessibility to high-throughput single cell transcriptomics.
Benefits of sample fixation for single cell studies
The scope and scale of single cell RNA sequencing studies is ever increasing, with researchers aiming to profile larger numbers of samples, corresponding to cohorts of patients or multiple treatment conditions or time points. Along with these ambitious experimental goals come additional considerations in sample acquisition and processing. Single Cell Gene Expression Flex creates new possibilities for high-throughput, batched analysis, and extends experimental capabilities to include fragile or dying cells, samples from multiple collection sites, and samples where cell state may be in flux. This new method provides a solution to the multitude of challenges that researchers have faced in trying to reach new depths in their studies.
"For projects including long-distance transportation of samples, cells or tissues may suffer the loss of viability from physical impact during transport or improper storage conditions. In some cases, sample preservation methods are required to allow more flexible experimental designs; specifically, it can help to store samples collected from different experimental conditions or time points and enable them to be consolidated. Besides, researchers may also be interested in specific biological states that in some tissues may become altered as specific pathways can be activated by in vitro processing." (9)
Preserving fragile and/or dynamic biology
By fixing samples at the point of collection, we can lock in biology that remains elusive with standard single cell transcriptomics. Fragile cells might not respond well to other storage methods, where something as routine as freeze-thaw cycles can cause damage. Along with storage, transportation to a lab for processing is also a potential danger to sample integrity. The resulting loss of some cell types or an increase in dead or dying cells can skew the results of your analysis, hiding the true heterogeneity of the sample in question. In a similar vein, studies that involve samples undergoing rapid change (e.g., drug screens with short time points, stimulating immune cells, etc.) would benefit from efficient fixation that preserves dynamic cell states, removing the urgency in processing.
Improving access to more samples
The size and overall organization of your study might lend itself well to fixed RNA profiling. Large experiments, in which you may be collecting numerous time points or testing under multiple conditions for mouse studies, cell lines, or organoids, can be overwhelming and labor-intensive if you are using protocols that require all of your samples to be processed at once. Fixing cells as they are collected can reduce the stress of these large-scale experiments, allowing you to process samples when it’s convenient. Perhaps you are conducting a study in which your samples are coming from different geographical locations and need to be shipped to a central lab for processing. Fixation locks in cell state and preserves sample quality for storage and shipment, increasing access to more samples without compromising integrity.
Accommodating day-to-day operations
How does your lab operate on a daily basis? If you are working with a core lab or central facility, their hours may be limited, restricting the times when fresh samples can be accepted. This could be of particular importance for situations in which your samples are ready for collection at unpredictable times of day or night. On the other hand, even within your own lab, you may be concerned about consistency between users or just from day to day. With fixed RNA-seq, you can store your samples for batch processing in a single run, minimizing variation.
Neutralizing safety concerns
Stepping back from these more operational concerns around throughput and sample management and viability, fixed RNA profiling can even improve researcher safety. If your work includes samples that contain infectious agents or pathogens, it may be inconvenient to perform all the processing for your single cell study in a BSL-3 lab. By fixing your samples, you can neutralize infectious agents. Following proper validation of this step, samples may no longer need to be handled in a BSL-3 lab, lending more freedom to your benchwork.
A familiar, easy-to-use workflow
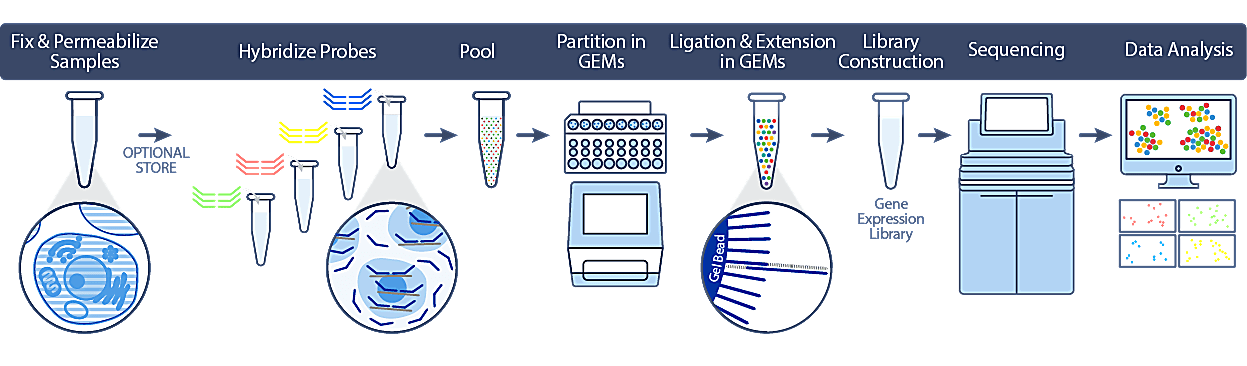
Single Cell Gene Expression Flex utilizes PFA fixation to preserve biology at the point of sample collection, streamlining workflows and improving access to more samples without time constraints. Leveraging our trusted Next GEM technology, we’ve developed a protocol that will be familiar to those who have used our Chromium Single Cell Gene Expression assays in the past, and easy to use even for those new to single cell transcriptomics.
This robust workflow includes optimized, demonstrated protocols for diverse sample types, including cell lines, primary cells, and dissociated tissue containing fragile cells.
As highlighted above, cells or nuclei are fixed and permeabilized at the point of collection, after which they can be safely stored and transported without compromising sample integrity or downstream data quality. When convenient, samples are hybridized to probe sets (covering over 18,000 genes) and can be processed individually using the single sample workflow, or pooled with up to 16 samples in a single lane of a Chromium chip using the multiplex workflow.
At this point, the workflow closely mirrors our 3’ and 5’ gene expression protocols, with single cell partitioning on our Chromium iX/X instrument. During this process, the probe sets are ligated and extended to incorporate unique barcodes to maintain cell identity. Following library construction and sequencing, analysis can be performed using 10x Genomics Cell Ranger and Loupe Browser software tools.
To fix or not to fix?
As with any new study, it’s critical to think through your experimental goals and how they affect the overall setup and, crucially, which assay(s) will help you reach those goals. For single cell analysis of genes, sample type, throughput capacity, and specific analytes will all influence this decision.
Species
Single Cell Gene Expression Flex is compatible with human and, soon, mouse samples. For other species, the Single Cell Gene Expression assay is the best option.
Throughput
If you are working with a large number of samples and would prefer to batch or multiplex them, two options are available: Single Cell Gene Expression Flex or Single Cell Gene Expression HT with 3’ CellPlex. Single Cell Gene Expression Flex enables sample multiplexing through unique barcodes that are built into the probe sets, while Single Cell Gene Expression HT with 3’ CellPlex labels pools of samples with species-agnostic tags. Similarly, for identifying rare cells or developing in-depth cell atlases, both of these assays can help you maximize cell throughput for a given sample.
Analytes
Multiomic analysis can add incredible detail to single cell experiments, and each of our single cell gene expression assays provide options for simultaneous measurement of multiple analytes. Transcriptome-wide gene expression can be paired with cell surface protein expression using the Single Cell Gene Expression kit or single sample workflow for Single Cell Gene Expression Flex. (View our Compatible Products page to find TotalSeq B oligo-conjugated antibodies from BioLegend for more information on antibody panels.)
If you'd like to access other multiomic insights, the following assays provide excellent opportunities for exploring more analytes, but are not compatible with PFA samples. For delving into immunology questions, Single Cell Immune Profiling provides the ability to analyze full-length, paired B- or T-cell receptors, surface protein expression, antigen specificity, and gene expression from a single cell. Our Single Cell CRISPR Screening assay allows you to directly assess specific gene edits or knockdowns at the same time as the resulting gene expression profile (and, soon, cell surface proteins too).
Innovative tools, transformative insights
As research continues to move forward in a quest to improve human health, we see new opportunities arise to overcome the challenges of larger and more complex studies. Single Cell Gene Expression Flex was developed to enhance access to more samples, preserving fragile biology and overcoming logistical challenges that have prevented such studies from moving forward. With these critical advancements in single cell technologies, scientists can investigate the molecular changes that influence physiology and pathology, transforming the way we understand disease progression and treatment.
We invite you to learn more about Single Cell Gene Expression Flex and other innovations for your molecular biology toolkit at our upcoming live webinar series. Additionally, you can start exploring how this new assay fits into your experimental goals by viewing data and workflow details.
References:
- Aldridge S and Teichmann SA. Single cell transcriptomics comes of age. Nat Commun 11: 4307 (2020). doi: 10.1038/s41467-020-18158-5
- Tang F, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6: 377–382 (2009). doi: 10.1038/nmeth.1315
- Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 26: 842–844 (2020). doi: 10.1038/s41591-020-0901-9
- Kim D, et al. Targeted therapy guided by single-cell transcriptomic analysis in drug-induced hypersensitivity syndrome: a case report. Nat Med 26: 236–243 (2020). doi: 10.1038/s41591-019-0733-7
- Stewart CA, et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer 1: 423–436 (2020). doi:s43018-019-0020-z
- Lynn RC, et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature 576: 293–300 (2019). doi: 10.1038/s41586-019-1805-z
- Smajić S, et al. Single-cell sequencing of human midbrain reveals glial activation and a Parkinson-specific neuronal state. Brain 2021. doi: 10.1093/brain/awab446
- De Miguel Z, et al. Exercise plasma boosts memory and dampens brain inflammation via clusterin. Nature 600: 494–499 (2021). doi: 10.1038/s41586-021-04183-x
- Wang X and Yu L. The effect of methanol fixation on single-cell RNA sequencing data. BMC Genomics 22: 420 (2021). doi: 10.1186/s12864-021-07744-6
About the author:

