The gold standard, transformed: Taking single cell RNA-seq further than ever before
Single cell technology has had a tremendous impact on the way scientists approach research into human health and disease—with far-reaching implications for studies into the effectiveness of CAR T-cell therapy for blood cancers (1), mechanisms of cognitive dysfunction (2), and treatments for diabetic kidney disease (3), just to name a few. Over the past decade, single cell RNA sequencing has been the most commonly used profiling technique (4), and yet, gaps in our capabilities have remained.
By and large, the use of single cell transcriptomics has come with logistical hurdles, most notably sample acquisition and processing. Working with fresh cells can be challenging and necessitates prompt sample preparation. Furthermore, accessing certain sample types can bring additional difficulties, with low-quality samples yielding damaged RNA, whole tissues requiring expertise in sample collection and preparation, and biobanked tissue lying out of reach without effective methods to extract intact cells or nuclei for analysis. If these obstacles persist, we risk losing out on critical data and novel insights. What does the way forward look like?
Improving access across sample types
For as much as single cell gene expression profiling kicked off a fundamental shift in biological research, this technology is now seeing its own transformation as we advance the chemistry to take scRNA-seq further than ever before. Chromium Single Cell Gene Expression Flex is a powerful probe-based assay that uses simple and ubiquitous fixation conditions to enable storage and transport of samples, without losing data quality or compromising sample integrity.
The ability to fix tissue prior to dissociating into single cells fills in the gaps of sample access, allowing you to preserve fragile biology at the point of collection, batch and run assays on your schedule, and perform whole transcriptome profiling on fresh or fixed samples, even FFPE. Additionally, the probe-based chemistry maximizes sensitivity, with three probe pairs per gene, and high compatibility with degraded RNA, as only a 50 bp footprint is required.
Note: You may already be familiar with Gene Expression Flex—it was first made available as Chromium Single Cell Fixed RNA Profiling (FRP). We previously discussed the assay and detailed the advantages of its unique protocol here. All of these benefits still apply, but we’ve updated the name to better reflect the incredible flexibility you gain in sample compatibility, experimental setup, and workflow logistics.
Gene Expression Flex vs. a tried-and-true standard
If you’ve run up against the traditional roadblocks in designing your single cell experiments, or had samples fail when using other methods, you understand the frustrations that can come along with this research. So let's take a look at how Gene Expression Flex can address these issues of accessibility and performance in a real-world example.
We’ve long held high our Chromium Single Cell kits to high standards, developing robust protocols that yield highly sensitive, reproducible results. Researchers at the Fred Hutch Innovation Lab recently ran a side-by-side comparison between our Chromium Single Cell Gene Expression 3’ kit and Gene Expression Flex. Their team used human peripheral blood mononuclear cells (PBMCs) for both assays. As per the respective protocols, they ran the 3’ samples fresh and fixed the Flex samples. Furthermore, they used an optional freezing protocol for the Flex kit, wherein fixed samples were frozen overnight, then thawed before running the assay.
They found that cell recovery for Gene Expression Flex exceeded that of the 3’ kit, and noted the added benefit of being able to re-freeze leftover cells—before or after probe hybridization—for use in future experiments. After sequencing all libraries and performing data analysis, the researchers observed that Flex was approximately twice as sensitive as our 3’ assay, with high reproducibility between replicates.
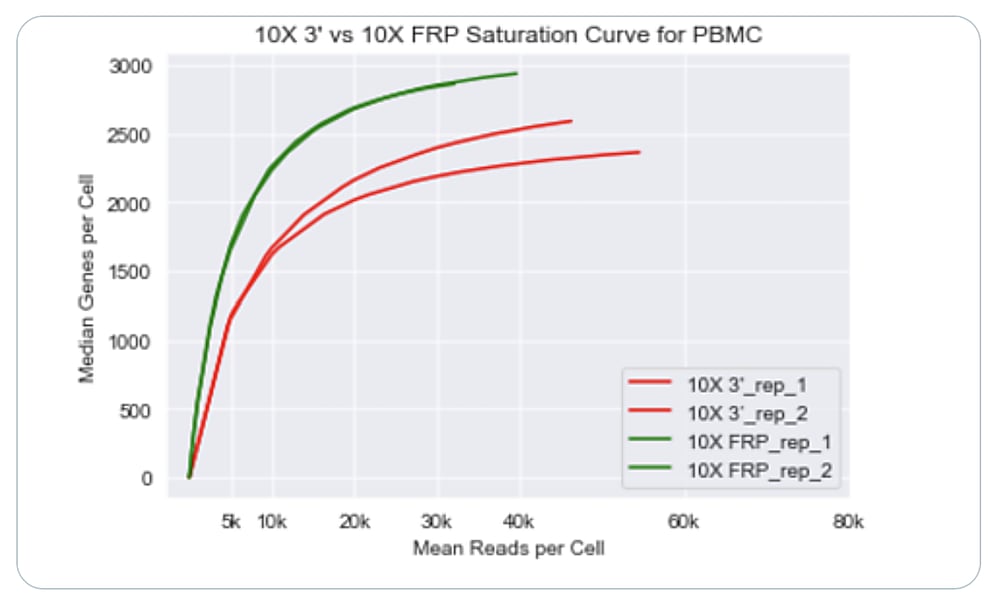
Visualization of each single cell dataset revealed cellular composition was maintained through both assays, with only minimal differences in the UMAP clustering between them.
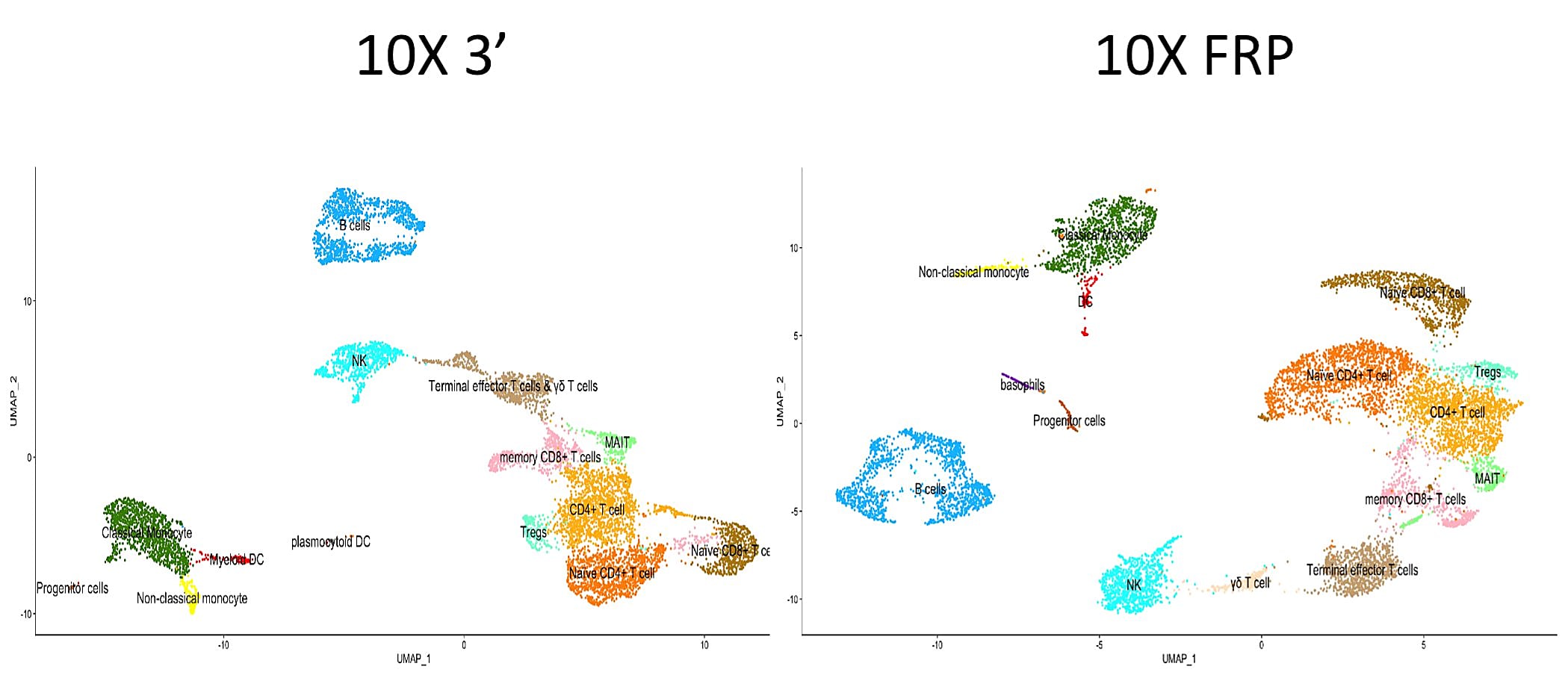
Additional data and details on experimental setup can be found in this thread from the Fred Hutch Innovation Lab.
Setting a new standard for single cell transcriptomics
The superior sensitivity and expanded sample access that Chromium Single Cell Gene Expression Flex brings to your bench translates into more opportunities to uncover biological insights. How well can you detect low-expressing genes? What about transcription factors? What rare cell types have gone unrepresented in earlier research? The data shared by the team at the Fred Hutch Innovation Lab echoes our own findings, demonstrating our highest scRNA-seq performance with Gene Expression Flex. We observe this same level of sensitivity across a wide range of sample types and under varied storage conditions. Usable reads and gene complexity remain comparable post-fixation for up to six months, and longer-term studies are currently underway.
As for archived tissue, our R&D team has worked hard to advance support for compatibility with FFPE. Now, a robust single cell protocol is available for those invaluable samples, enabling dissociation of FFPE tissue for use with Single Cell Gene Expression Flex. You can download the Demonstrated Protocol, complete with a reagent list and current information on validation across a wide range of tissue types.
The strength and versatility of Gene Expression Flex can empower single cell studies in remarkable new ways, pushing your research further than ever before. We can’t wait to see how you move forward from here, and if the past ten years are any indication, a wealth of information is waiting to be explored.
Learn more about Chromium Single Cell Gene Expression Flex here.
References:
- Dong H, et al. Memory-like NK cells armed with a neoepitope-specific CAR exhibit potent activity against NPM1 mutated acute myeloid leukemia. Proc Natl Acad Sci USA 119: e2122379119 (2022). doi: 10.1073/pnas.2122379119
- Fu Y, et al. Single-nucleus RNA sequencing reveals the shared mechanisms inducing cognitive impairment between COVID-19 and Alzheimer’s disease. Front Immunol 13: 967356 (2022). doi: 10.3389/fimmu.2022.967356
- Wu H, et al. Mapping the single-cell transcriptomic response of murine diabetic kidney disease to therapies. Cell Metab 34: 1064–1078.e6 (2022). doi: 10.1016/j.cmet.2022.05.010
- Perkel, J M. Single-cell analysis enters the multiomics age. Nature (2021)
