Cluster mode is one of four primary ways of running Cell Ranger. To learn about the other approaches, go to the computing options page. Cell Ranger can be run in cluster mode using Slurm to execute the stages on multiple nodes via batch scheduling. This allows highly parallelizable stages to use hundreds or thousands of cores concurrently, dramatically reducing time to results.
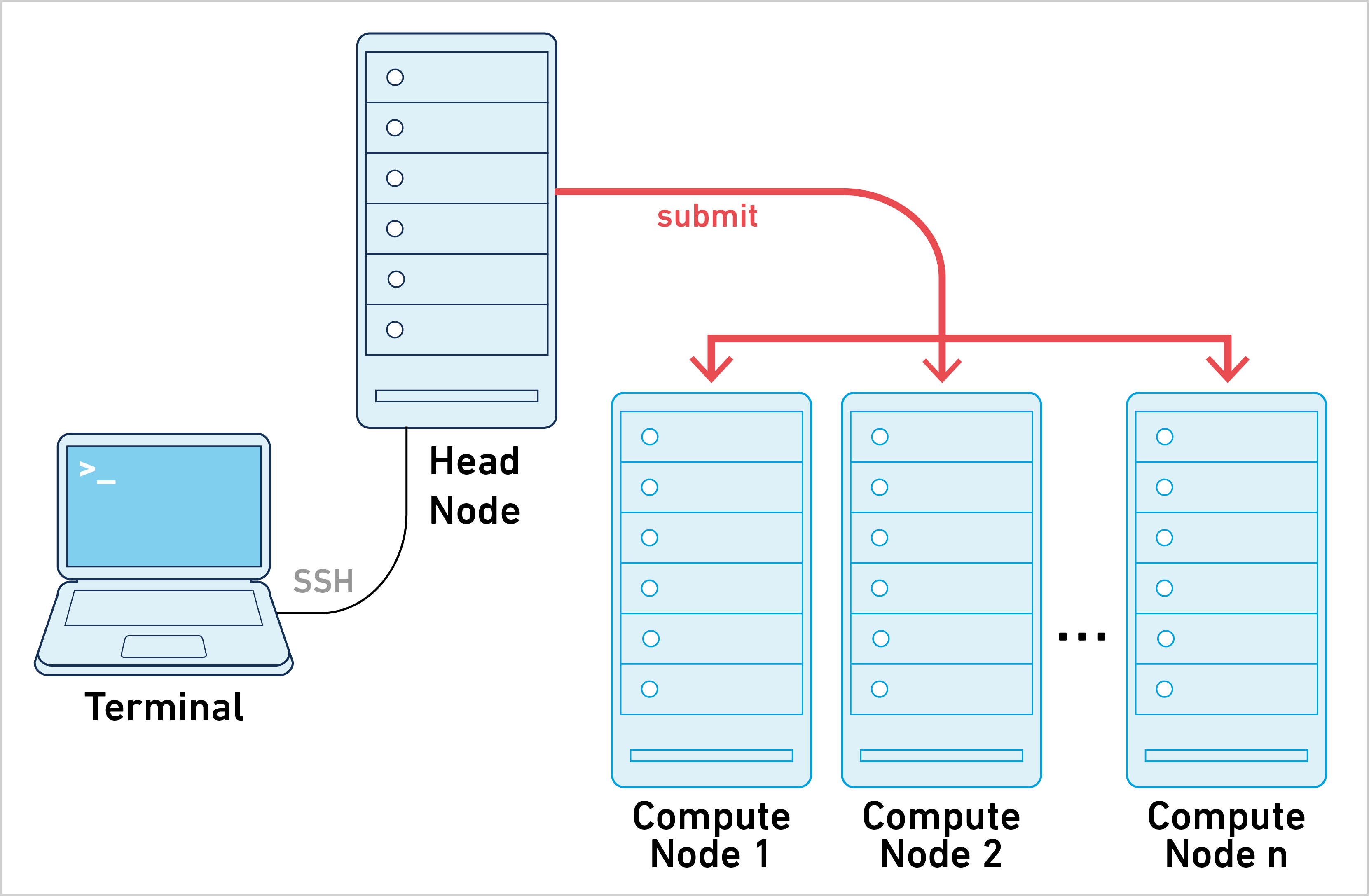
Running pipelines in cluster mode requires the following: 1. Cell Ranger is installed in the same location on all nodes of the cluster. For example, /opt/cellranger-7.1.0 or /net/apps/cellranger-7.1.0 2. Cell Ranger pipelines are run on a shared file system accessible to all nodes of the cluster. NFS-mounted directories are the most common solution for this requirement. 3. The cluster accepts both single-core and multithreaded (shared-memory) jobs.
Installing the Cell Ranger software on a cluster is identical to installation on a local server. After confirming that the cellranger commands can run in single server mode, configure the job submission template that Cell Ranger uses to submit jobs to the cluster. Assuming Cell Ranger is installed to /opt/cellranger-7.1.0, the process is as follows.
Step 1. Navigate to the Martian runtime's jobmanagers/ directory that contains example jobmanager templates.
# command to change directory
cd /opt/cellranger-7.1.0/external/martian/jobmanagers
# command to list files
tree -L 2
.
├── config.json
├── lsf.template.example
├── pbspro.template.example
├── retry.json
├── sge_queue.py
├── sge.template.example
├── slurm_queue.py
├── slurm.template.example
└── torque.template.example
Step 2. Make a copy of the cluster's example Slurm template to slurm.template in the jobmanagers/ directory.
# command to copy file
cp -v slurm.template.example slurm.template
# Output looks like this:
`slurm.template.example' -> `slurm.template'
List the files again to check if the copy command worked:
# command to list files
tree -L 2
.
├── config.json
├── lsf.template.example
├── pbspro.template.example
├── retry.json
├── sge_queue.py
├── sge.template.example
├── slurm_queue.py
├── slurm.template
├── slurm.template.example
└── torque.template.example
The job submission templates contain several special variables that are substituted by the Martian runtime when each stage is submitted. Specifically, the following variables are expanded when a pipeline is submitting jobs to the cluster:
| Variable | Must be present? | Description |
|---|---|---|
__MRO_JOB_NAME__ | Yes | Job name composed of the sample ID and stage being executed |
__MRO_THREADS__ | No | Number of threads required by the stage |
__MRO_MEM_GB__ __MRO_MEM_MB__ | No | Amount of memory (in GB or MB) required by the stage |
__MRO_MEM_GB_PER_THREAD__ __MRO_MEM_MB_PER_THREAD__ | No | Amount of memory (in GB or MB) required per thread in multithreaded stages. |
__MRO_STDOUT__ __MRO_STDERR__ | Yes | Paths to the _stdout and _stderr metadata files for the stage |
__MRO_CMD__ | Yes | Bourne shell command to run the stage code |
It is critical that the special variables listed as required are present in the final template you create. In most cases, the templates will not need modification. The exception to this rule is
#SBATCH --ntasks=1 --cpus-per-task=__MRO_THREADS__
### Alternatively: --ntasks-per-node=__MRO_THREADS__
Consult with your cluster administrators or help desk to find the combination that works best for single-node, multi-threaded applications on your system.
Depending on which job scheduler you have, select an option below.
SGE
For SGE cluster, you MUST replace <pe_name> within the example template to reflect the name of the cluster's multithreaded parallel environment. To view a list of the cluster's parallel environments, use the qconf -spl command.
The most common modifications to the job submission template include adding additional lines to specify:
- The research group's queue. For example,
#$ -q smith.q - The account to which jobs will be charged. For example,
#$ -A smith_lab
LSF
The most common modifications to the job submission template include adding additional lines to specify:
- Your research group's queue. For example,
#BSUB -q smith_queue - The account to which your jobs will be charged. For example,
#BSUB -P smith_lab
To run a Cell Ranger pipeline in cluster mode, add
the --jobmode=slurm, --jobmode=sge, or --jobmode=lsf command line option when using
the cellranger commands. It is also possible to use --jobmode=<PATH>,
where <PATH> is the full path to the cluster template file.
To validate that cluster mode is properly configured, follow the same validation instructions given for cellranger in the Installation page, but add --jobmode=slurm, --jobmode=sge, or --jobmode=lsf.
cellranger mkfastq --run=./tiny-bcl --samplesheet=./tiny-sheet.csv --jobmode=slurm
Martian Runtime - 7.1.0
Running preflight checks (please wait)...
2016-09-13 12:00:00 [runtime] (ready) ID.HAWT7ADXX.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
2016-09-13 12:00:00 [runtime] (split_complete) ID.HAWT7ADXX.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET
...
Once the preflight checks are finished, check the job queue to see the stages queuing up (e.g., using the squeue command for Slurm).
In the event of a pipeline failure, an error message is displayed.
[error] Pipestance failed. Please see the log at:
HAWT7ADXX/MAKE_FASTQS_CS/MAKE_FASTQS/MAKE_FASTQS_PREFLIGHT/fork0/chnk0/_errors
Saving diagnostics to HAWT7ADXX/HAWT7ADXX.debug.tgz
For assistance, upload this file to 10x by running:
uploadto10x <your_email> HAWT7ADXX/HAWT7ADXX.debug.tgz
The _errors file contains a jobcmd error:
cat HAWT7ADXX/MAKE_FASTQS_CS/MAKE_FASTQS/MAKE_FASTQS_PREFLIGHT/fork0/chnk0/_errors
jobcmd error:
exit status 1
The most likely reason for this failure is an invalid job submission template. This occurs when the job submission via sbatch, qsub, or bsub commands failed.
The "tiny" dataset does not stress the cluster; it may be worthwhile to follow up with a more realistic test using one of our sample datasets.
There are two subtle variants of running cellranger pipelines in cluster mode, each with its pitfalls. Check with your cluster administrator to see which approach is compatible with your institution's setup.
-
Run
cellrangerwith--jobmode=slurmon the head node. Cluster mode was originally designed for this use case. However, this approach leavesmrpandmrjobrunning on the head node for the duration of the pipeline, and some clusters impose time limits to prevent long-running processes. -
Use a job script to submit a
cellrangercommand with--jobmode=slurm: With this approach,mrpandmrjobrun on a cluster mode. However, the cluster must allow jobs to be submitted from a compute node to make this viable.
When cellranger is run in cluster mode, a single library analysis is partitioned into hundreds and potentially thousands of smaller jobs. The underlying Martian pipeline framework launches each stage job using the sbatch, qsub, or bsub commands when running on the Slurm, SGE, or LSF cluster, respectively. As stage jobs are queued, launched, and completed, the pipeline framework tracks their status using the metadata files that each stage maintains in the pipeline output directory.
Like single server pipelines, cluster-mode pipelines can be restarted after a failure. They maintain the same order of execution for dependent stages of the pipeline. All executed stage code is identical to single server mode, and the quantitative results are identical to the limit of each stage's reproducibility.
cellranger commands have exited.In addition, the Cell Ranger UI can still be used with cluster mode. Because the Martian pipeline framework runs on the node from which the command was issued, the UI will also run from that node.
Stages in the Cell Ranger pipelines each request a specific number of cores and memory to aid with resource management. These values are used to prevent oversubscription of the computing system when running pipelines in single server mode. The way CPU and memory requests are handled in cluster mode is defined by the following:
- How the
__MRO_THREADS__and__MRO_MEM_GB__variables are used within the job template. - How your specific cluster's job manager schedules resources.
Depending on which job scheduler you have, select an option below.
SGE supports requesting memory via the mem_free resource natively, although your cluster may have another mechanism for requesting memory. To pass each stage's memory request through to SGE, add line to your sge.template that requests mem_free, h_vmem, h_rss, or the custom memory resource defined by your cluster:
cat sge.template
#$ -N __MRO_JOB_NAME__
#$ -V
#$ -pe threads __MRO_THREADS__
#$ -l mem_free=__MRO_MEM_GB__G
#$ -cwd
#$ -o __MRO_STDOUT__
#$ -e __MRO_STDERR__
__MRO_CMD__
The h_vmem (virtual memory) and mem_free/h_rss (physical memory) represent two different quantities and Cell Ranger stages' __MRO_MEM_GB__ requests are expressed as physical memory. Using h_vmem in your job template may cause certain stages to be unduly killed if their virtual memory consumption is substantially larger than their physical memory consumption. It follows that we do not recommend using h_vmem.
For clusters whose job managers do not support memory requests, it is possible to request memory in the form of cores via the --mempercore command line option. This option scales up the number of threads requested via the __MRO_THREADS__ variable according to how much memory a stage requires.
For example, given a cluster with nodes that have 16 cores and 128 GB of memory (8 GB per core), the following pipeline invocation command:
cellranger mkfastq --run=./tiny-bcl --samplesheet=./tiny-sheet.csv --jobmode=slurm --mempercore=8
will issue the following resource requests:
- A stage that requires 1 thread and less than 8 GB of memory will pass
__MRO_THREADS__of1to the job template. - A stage that requires 1 thread and 12 GB of memory will pass
__MRO_THREADS__of2to the job template because (12 GB) / (8 GB/core) = 2 cores. - A stage that requires 2 threads and less than 16 GB of memory will pass
__MRO_THREADS__of2to the job template. - A stage that requires 2 threads and 40 GB of memory will pass
__MRO_THREADS__of5to the job template because (40 GB) / (8 GB/core) = 5 cores.
As the final bullet point illustrates, this mode can result in wasted CPU cycles and is only provided for clusters that cannot allocate memory as an independent resource.
Every cluster configuration is different. If you are unsure of how your cluster resource management is configured, contact your cluster administrator or help desk.
LSF supports job memory requests through the -M and -R [mem=...] options, but these requests generally must be expressed in MB,
not GB. As such, your LSF job template should use
the __MRO_MEM_MB__ variable rather than __MRO_MEM_GB__. For example,
cat bsub.template
#BSUB -J __MRO_JOB_NAME__
#BSUB -n __MRO_THREADS__
#BSUB -o __MRO_STDOUT__
#BSUB -e __MRO_STDERR__
#BSUB -R "rusage[mem=__MRO_MEM_MB__]"
#BSUB -R span[hosts=1]
__MRO_CMD__
For clusters whose job managers do not support memory requests, it is possible to request memory in the form of cores via the --mempercore command line option. This option scales up the number of threads requested via the __MRO_THREADS__ variable according to how much memory a stage requires.
For example, given a cluster with nodes that have 16 cores and 128 GB of memory (8 GB per core), the following pipeline invocation command:
cellranger mkfastq --run=./tiny-bcl --samplesheet=./tiny-sheet.csv --jobmode=slurm --mempercore=8
will issue the following resource requests:
- A stage that requires 1 thread and less than 8 GB of memory will pass
__MRO_THREADS__of1to the job template. - A stage that requires 1 thread and 12 GB of memory will pass
__MRO_THREADS__of2to the job template because (12 GB) / (8 GB/core) = 2 cores. - A stage that requires 2 threads and less than 16 GB of memory will pass
__MRO_THREADS__of2to the job template. - A stage that requires 2 threads and 40 GB of memory will pass
__MRO_THREADS__of5to the job template because (40 GB) / (8 GB/core) = 5 cores.
As the final bullet point illustrates, this mode can result in wasted CPU cycles and is only provided for clusters that cannot allocate memory as an independent resource.
Every cluster configuration is different. If you are unsure of how your cluster resource management is configured, contact your cluster administrator or help desk.
Each stage requests several threads and maximum free memory. These values are hardcoded into each stage and determined empirically by looking at in-house data runs, as well as reports from our customers. You may find that on your data, certain stages do not require as much memory as requested, or may require more memory than our defaults. The latter is more serious, as clusters may impose strict memory limits, and kill a job if those limits are exceeded.
You can override the defaults of a stage by supplying an override.json file, and specifying this file as the --override argument to your pipeline. Here is an example of an override JSON file to Cell Ranger, which overrides the memory requests of the count pipelines:
{
"SC_RNA_COUNTER_CS.SC_MULTI_CORE.MULTI_GEM_WELL_PROCESSOR.COUNT_GEM_WELL_PROCESSOR._BASIC_SC_RNA_COUNTER._MATRIX_COMPUTER.MAKE_SHARD": {
"chunk.mem_gb": 16,
"chunk.threads": 2
}
}
This configuration will increase the memory and threads requested (originally 6GB and 1 thread).
To run a pipeline with the above configuration, supply the JSON file as the --override parameter:
cellranger count --id=sample ... --override=./override.json
Overrides apply to pipelines executed both on the cluster and in local mode, but are likely most applicable to cluster mode users.
Some Cell Ranger pipeline stages are divided into hundreds of jobs. By default, the rate at which these jobs are submitted to the cluster is throttled to at most 64 at a time and at least 100 ms between each submission to avoid running into limits on clusters that impose quotas on the total number of pending jobs a user can submit.
If your cluster does not have these limits or is not shared with other users, you can control how the Martian pipeline runner sends job submissions to the cluster by using the --maxjobs and --jobinterval parameters.
To increase the cap on the number of concurrent jobs to 200, use the --maxjobs parameter:
cellranger count --id=sample ... --jobmode=slurm --maxjobs=200
You can also change the rate limit on how often the Martian pipeline runner sends submissions to the cluster. To add a five-second pause between job submissions, use the --jobinterval parameter:
cellranger count --id=sample ... --jobmode=slurm --jobinterval=5000
The job interval parameter is in milliseconds. The minimum allowable value is 1.