Exciting research is being done using the 10x Genomics Chromium Single Cell Gene Expression Solution. This guide outlines how to perform the analysis and highlights the results 10x Genomics assays and software produce using data from a recent Nature publication "Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell" (2019; doi: 10.1038/s41586-019-1154-y).
Intestinal tissue can repair itself after an injury such as irradiation. However, the molecular mechanisms that underlie the process are not fully understood. The LGR5+ crypt base columnar cells are thought to drive intestinal epithelium regeneration, but these cells are lost after injury, while regeneration still takes place.
To identify the cells responsible for the regeneration process, a group of researchers in Toronto used the 10x Genomics Chromium Single Cell 3’ Gene Expression Solution to profile mouse intestine cells, with and without irradiation.
This guide focuses on two samples: crypts (enriched from the whole epithelia) from normal and irradiated mice. See the figure below.
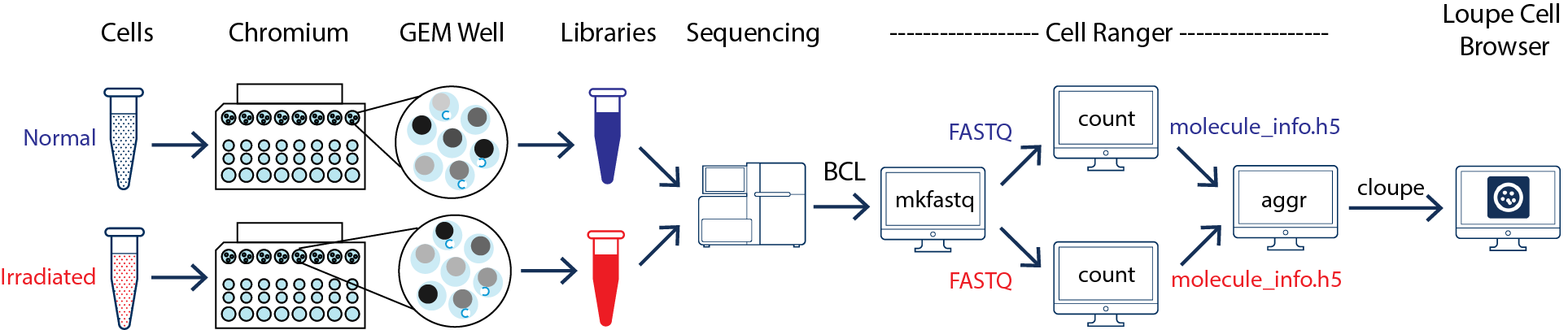
As illustrated above, the two samples were processed in two GEM wells on the 10x Chromium Controller. The two libraries were then prepared following the user guide and sequenced on a recommended Illumina sequencer.
The sequencer generates raw data in the base call (BCL) format, which contains sequencing data of all the libraries in the sequencing run. Use one of Illumina's demultiplexing pipelines to demultiplex BCL files into FASTQ files. If the sequencing provider already completed this step, the FASTQ files of each library can be directly used for data analysis.
For this guide, the Barcoded BAM files for the two samples was downloaded from the Data access tab of the Run Browser from these two Sequence Read Archive (SRA) pages:
The Barcoded BAM files are converted to FASTQ files with the 10x Genomics bamtofastq tool as below:
bamtofastq C05.bam.1 normal
bamtofastq C07.bam.1 irradiated
After successfully completing the bamtofastq, both normal and irradiated folders contain two subfolders with FASTQ files in them.
Once the FASTQ files for each sample are generated, the data analysis begins. The cellranger count pipeline can perform read alignment, UMI counting, and secondary analysis (dimensionality reduction, clustering, and visualization) for a single sample. The two samples shown in the figure above require running cellranger count for each sample separately.
Given that these are mouse samples, the pre-built mouse reference is used. After determining the path to the reference folder and FASTQ files, run cellranger count for the normal sample:
cd ./normal/
cellranger count --id=normal \
--transcriptome=/path/to/refdata-cellranger-mm10-3.0.0 \
--fastqs=./indepth_C05_MissingLibrary_1_HL5G3BBXX,./indepth_C05_MissingLibrary_1_HNNWNBBXX
Similarly, run cellranger count for the irradiated sample in a separate command:
cd ./irradiated/
cellranger count --id=irradiated \
--transcriptome=/path/to/refdata-cellranger-mm10-3.0.0 \
--fastqs=./indepth_C07_MissingLibrary_1_HL5G3BBXX,./indepth_C07_MissingLibrary_1_HNNWNBBXX
After successfully completing the pipeline, find the outs/ directory for each run to review many useful result files. Start by checking the results using the following files:
- The
web_summary.htmlcontains summary metrics and automated secondary analysis results. Click the ? in the top of each dashboard for more information on each metric. If there are any metrics with abnormal values, Cell Ranger automatically generates alerts to help you identify any potential issues. If you have specific questions, you can search our Q&A articles or contact support. - If the metrics look reasonable, load the
cloupe.cloupeinto Loupe Browser to visualize and explore the results. - For additional analysis, load the
filtered_feature_bc_matrixinto third-party tools, such as Seurat.
For the example data here, there were no alerts for both normal and irradiated samples. To identify the cells that contribute to the intestine regeneration, we need to compare the results for normal versus irradiated cells.
To compare two or more samples, use the cellranger aggr pipeline (also see the getting started tutorial for Running cellranger aggr) to aggregate outputs from multiple runs of cellranger count, normalize runs to the same effective sequencing depth, and then perform secondary analysis on the combined data.
In this example, there are two samples, so the cellranger aggr pipeline requires a libraries.csv file that looks like this:
library_id,molecule_h5
normal,/path/to/count/normal/outs/molecule_info.h5
irradiated,/path/to/count/irradiated/outs/molecule_info.h5
Here is an example of the cellranger aggr command:
cellranger aggr --id=aggr --csv=libraries.csv
Upon completion, this pipeline generates a set of outputs with the combined data from the two samples. Next, we can visualize and explore the results using the cloupe.cloupe file.
The combined data from two samples can be analyzed using Loupe Browser. Some key functions of the Loupe Browser are presented in this guide. For more detailed information, see the Loupe Browser tutorials.
To identify the cell type contributing to intestine regeneration, locate the major known cell types in the results. This step heavily relies on understanding the gene markers in the tissue of interest. There are also third-party computational tools developed for automated identification of cell types. For this guide, the typical method is used, which utilizes unsupervised clustering to identify the signature genes and associate them to known cell types based on the literature.
First, review the clustering results. The graph-based clustering results from the combined normal and irradiated samples showed 22 clusters. See the screenshot below. While not exactly the same as reported in the publication due mainly to software and version differences, the following sections of this guide illustrate some of the major results that were reproduced from the publication using Cell Ranger v3.1.0 pipelines.
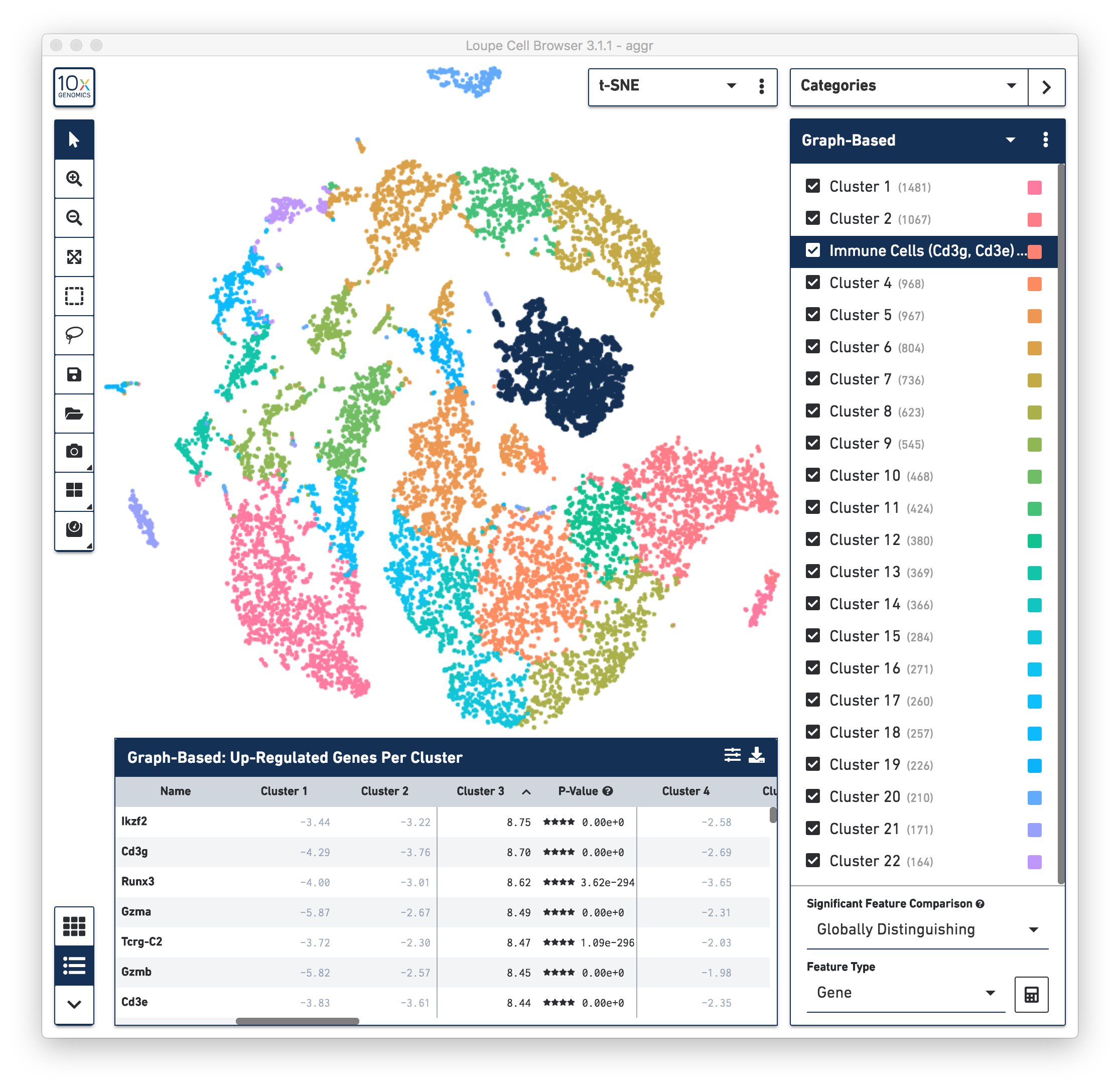
To identify the cell types of each cluster, use the differentially expressed genes shown in the data panel on the bottom. For example, in the above screenshot, see the top up-regulated genes in Cluster 3. Some of the genes are known immune cell markers (e.g., Cd3g and Cd3e), indicating that the cells in Cluster 3 are immune cells. Rename the Cluster 3 to "Immune Cells (Cd3g, Cd3e)".
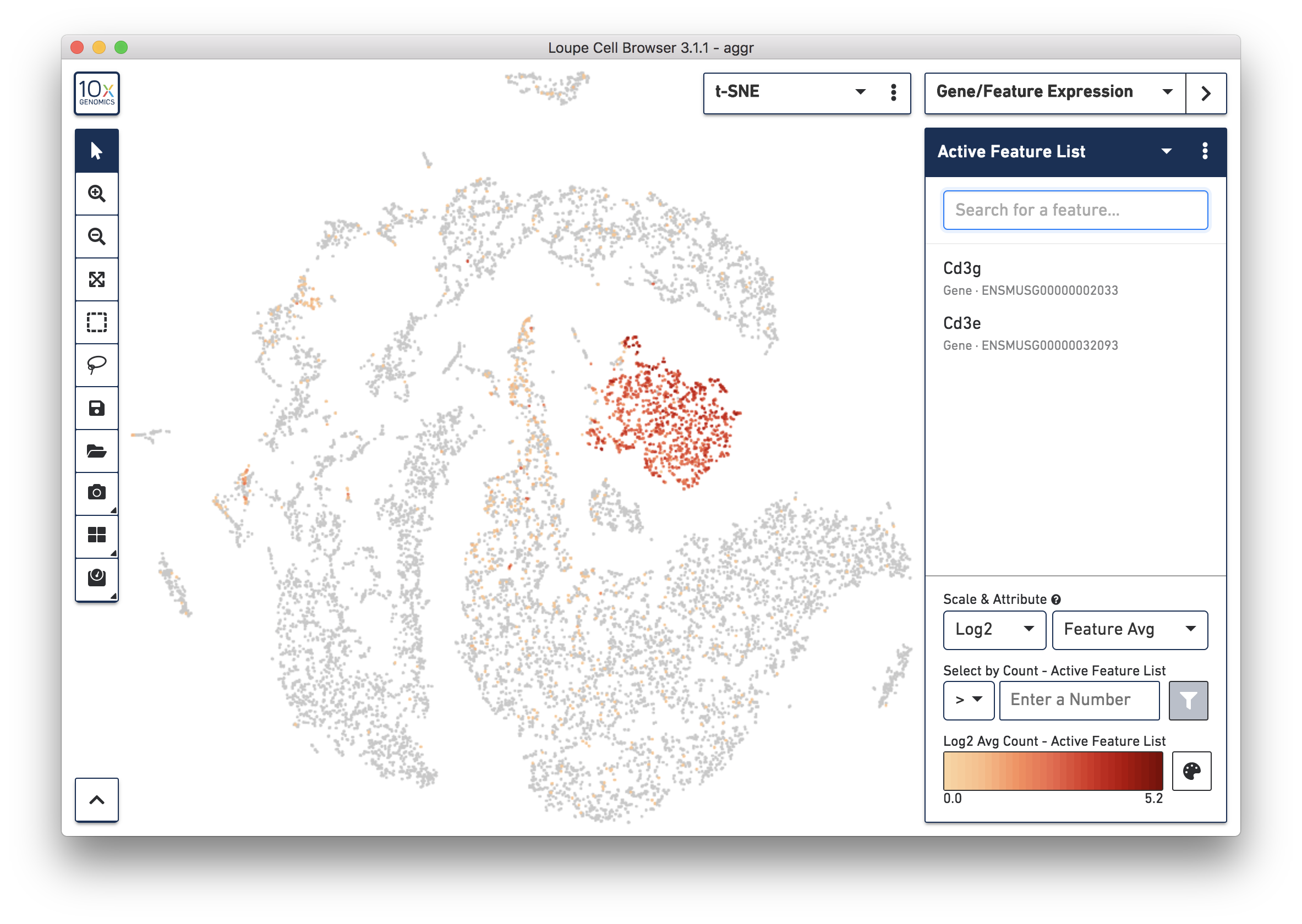
To further confirm that Cluster 3 represents immune cells, use the Gene/Feature Expression mode to view the expression level of Cd3g and Cd3e across the dataset. See the screenshot below. The cells expressing these immune cell markers are neatly clustered into Cluster 3, indicating that this distinct region of cells represents immune cells.
Similarly, identify the cell types of all the other clusters based on the top up-regulated genes and associate them with known cell types based on the literature. Depending on the complexity of the dataset and prior knowledge of the cell types, this process could be time-consuming.
Consistent with results reported in the publication, all but one cluster was assigned to known cell types. The cell types and marker genes identified in Loupe Browser are shown in the screenshot below.
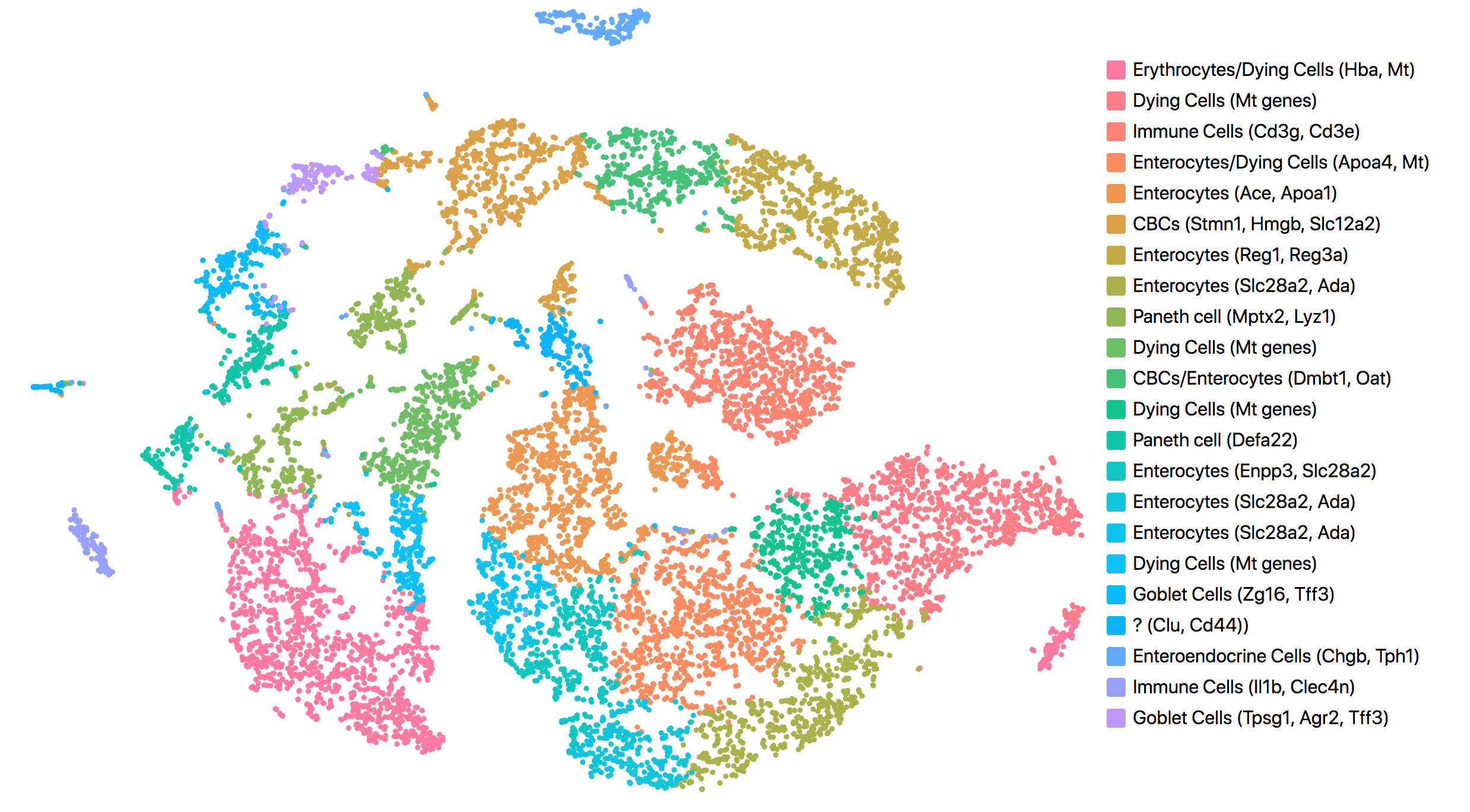
The most up-regulated gene in the unknown cell cluster is clusterin (Clu), which is consistent with the publication.
Next, continue using Loupe Browser to explore if the cluster of unknown cells is enriched or lost in the irradiated sample compared to the normal one. To see that more cells in this unknown cluster were from the irradiated sample, split the view by LibraryID category (see the screenshot below) and select only the unknown cluster.
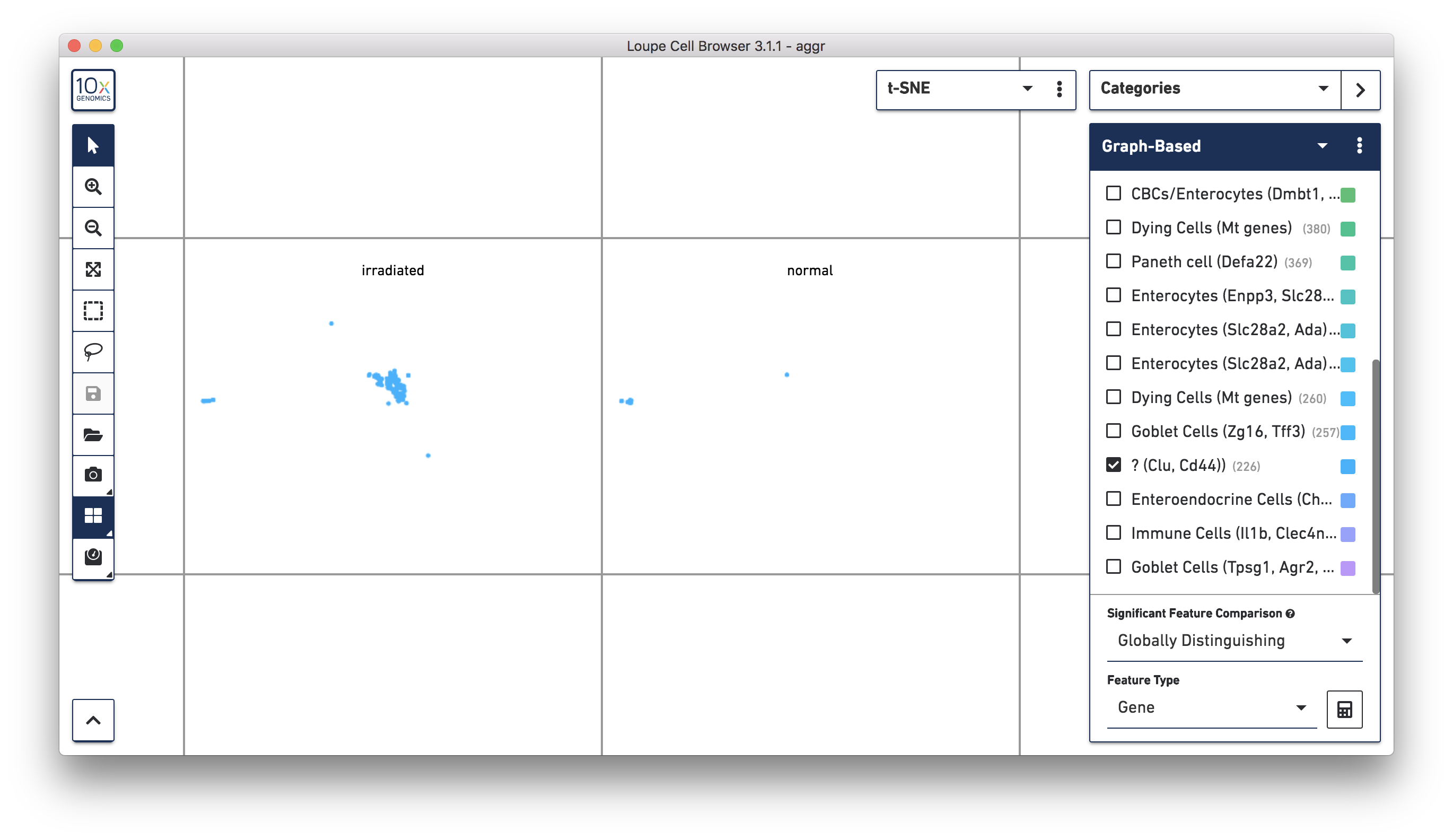
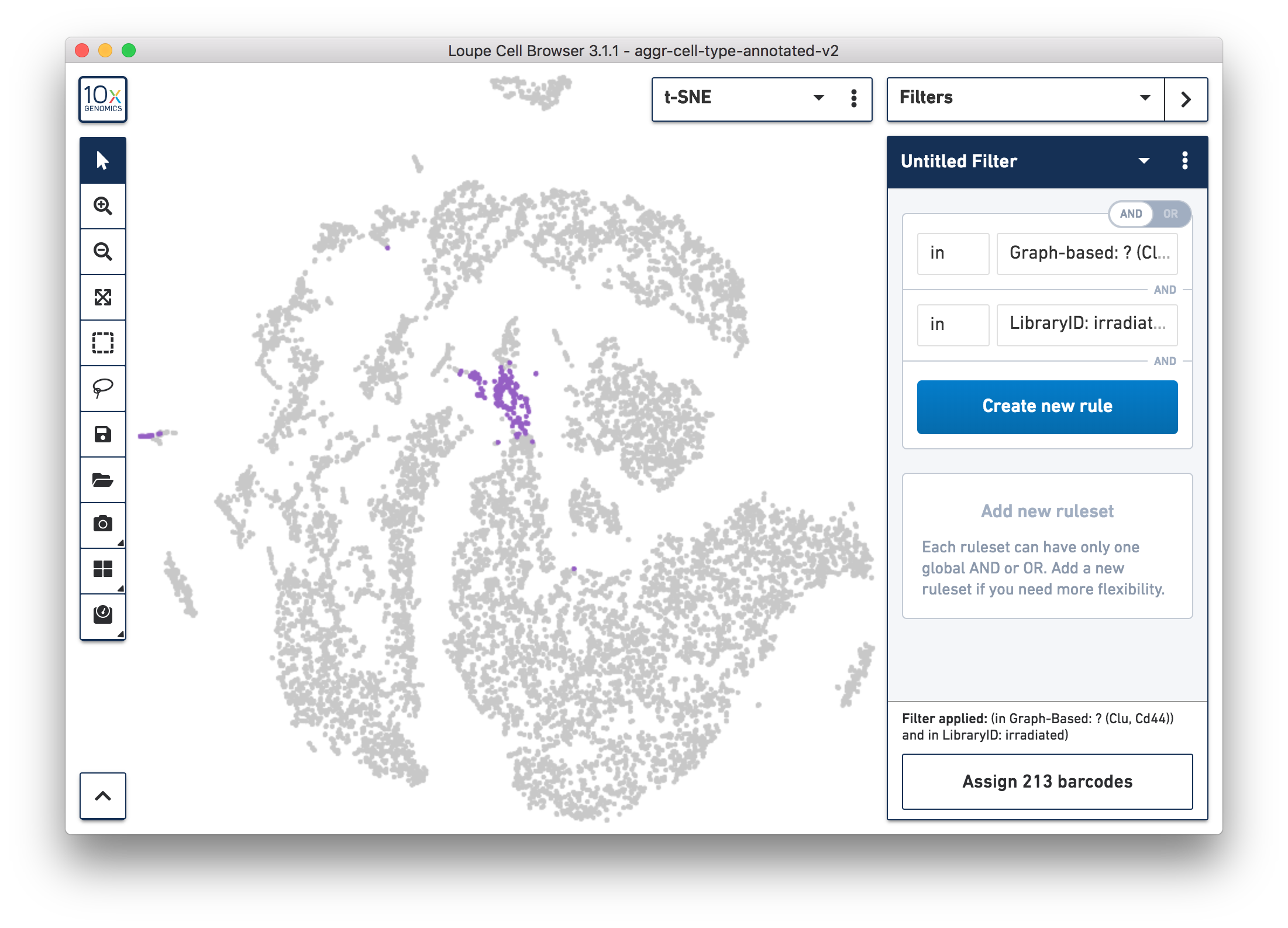
For more accurate numbers, use the Filters panel (see the screenshot below). 213 cell barcodes from the irradiated sample were found in this unknown cluster, while only 13 cells from the normal sample were in this cluster. This result is also similar to what was reported in the publication.
Following these initial findings, the authors performed additional experiments and validated that the Clu+ cell induced by injury is essential for intestine regeneration.
This was a quick tour of how to use the 10x Genomics Chromium Single Cell Gene Expression Solution and analysis tools to identify a novel cell type. Additional analyses can be done using 10x Genomics tools or third-party tools.
For example, in the results, several clusters of cells are enriched for mitochondrial (MT) genes. They can be excluded in the secondary analysis by following the instructions in this article: How can I exclude cells that show enrichment of MT genes from secondary data analysis?
Output data from Cell Ranger can be loaded into third-party tools to perform trajectory analysis, which can be used to explore the role of the Clu+ cells in the dynamic process of intestine regeneration.
To get started with your own single cell gene expression experiments, visit our gene expression support website for more details on workflow and software.