Cell Ranger v10.0 and later supports analyzing Flex v2 data with the cellranger multi pipeline. For general information on setting up and running the multi pipeline, visit the Cell Ranger multi pipeline page.
The cellranger multi pipeline relies on a specific multi config CSV structure for proper execution. Failure to comply with the required format (e.g., column headers, delimiters) will lead to parsing errors. Ensure that the multi config is saved in CSV format with the CSV extension.
- See the Cell Ranger Multi Config CSV page for a complete list of options for each section.
- To generate a multi config CSV template, run
cellranger multi-templateand see the usage instructions here.
Examples of multi config CSVs for the most common Flex v2 library combinations are provided below. If your specific library combination is not listed and you need assistance, contact support@10xgenomics.com.
When using cellranger multi to analyze Flex v2 plate-based workflows, keep in mind that one run of cellranger multi corresponds with one GEM Well. For example, in the following diagram, 96 sample barcodes are used (one plate), 24 sample barcodes are loaded per GEM Well, therefore four cellranger multi runs are required.
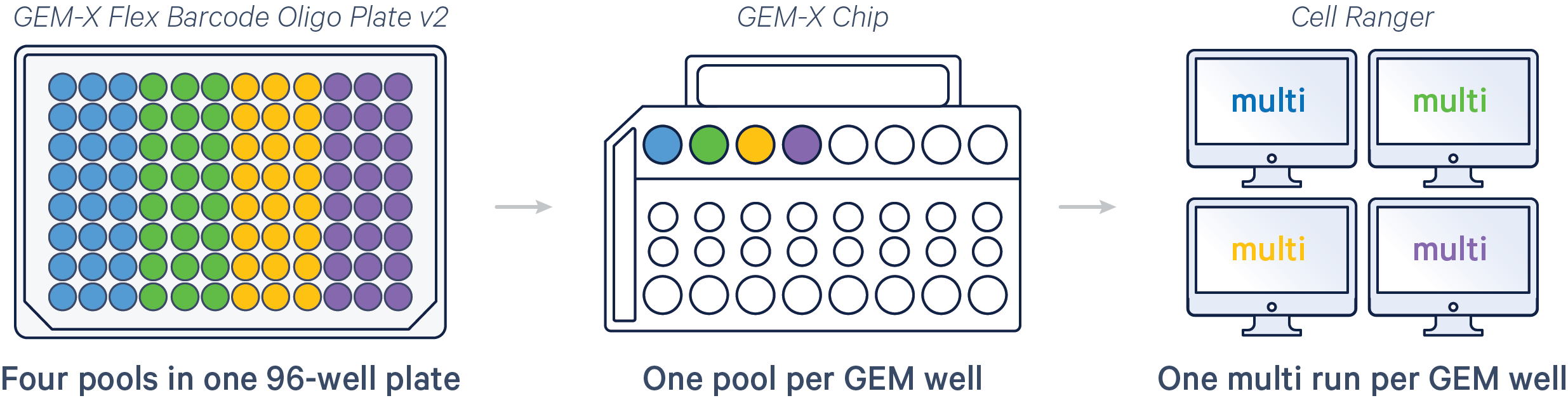
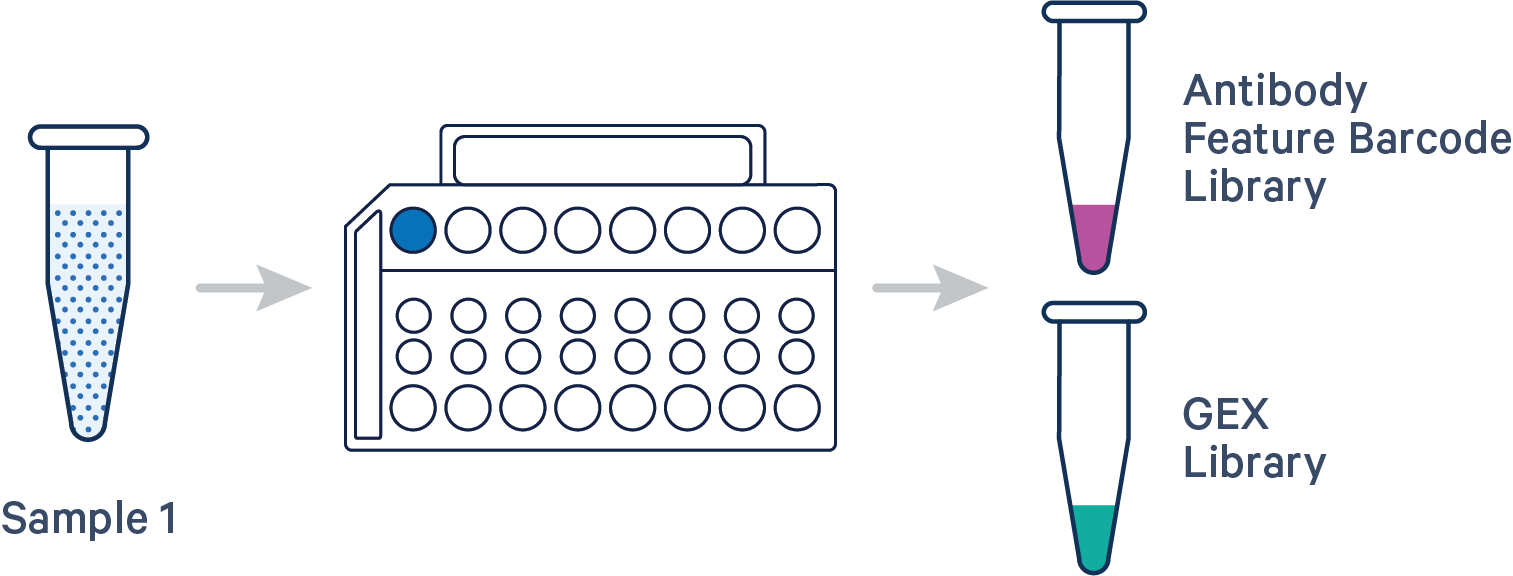
Here are a few example multi config CSVs for some common Flex v2 assay configurations, along with simplified diagrams for the corresponding experimental set up. Replace /path/to with the absolute path to your data, and customize the text according to the experiment's sample, library, and file names. Ensure that the multi config is saved in CSV format with the CSV extension.
Important: In the examples below, we set create-bam to "false" so Cell Ranger will not generate a BAM file. This setting is recommended for Flex libraries and will reduce both the total computation time for the pipestance and the size of the output directory.
The feature barcoding library is optional. If you do not have a feature barcoding library (i.e. Antibody or CRISPR), omit the [feature] section and include only the gene expression library information in the [libraries] section.
[gene-expression]
probe-set,/path/to/probe-set.csv #e.g., cellranger-x.y.z/probe_sets/Chromium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2024-A.csv
create-bam,false #do not generate BAM file
[libraries]
fastq_id,fastqs,feature_types
flex_gex,/path/to/fastqs,Gene Expression
flex_ab,/path/to/fastqs,Antibody Capture
[feature]
reference,/path/to/feature_reference.csv
The feature barcoding library is optional. If you do not have a feature barcoding library (i.e. Antibody or CRISPR), omit the [feature] section and include only the gene expression library information in the [libraries] section.
The probe_barcode_id is in the format of <plate_id>-<well_position>. Example templates can be downloaded here:

[gene-expression]
probe-set,/path/to/probe-set.csv #e.g., cellranger-x.y.z/probe_sets/Chromium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2024-A.csv
create-bam,false #do not generate BAM file
[libraries]
fastq_id,fastqs,feature_types
flex_gex,/path/to/fastqs,Gene Expression
flex_ab,/path/to/fastqs,Antibody Capture
[feature]
reference,/path/to/feature_reference.csv
[samples]
Sample_id,probe_barcode_ids
Sample1,A-A01
Sample2,A-B01
Sample3,A-C01
Sample4,A-D01
The feature barcoding library is optional. If you do not have a feature barcoding library (i.e. Antibody or CRISPR), omit the [feature] section and include only the gene expression library information in the [libraries] section.

[gene-expression]
probe-set,/path/to/probe-set.csv #e.g., cellranger-x.y.z/probe_sets/Chromium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2024-A.csv
create-bam,false #do not generate BAM file
[libraries]
fastq_id,fastqs,feature_types
flex_gex,/path/to/fastqs,Gene Expression
flex_ab,/path/to/fastqs,Antibody Capture
[feature]
reference,/path/to/feature_reference.csv
[samples]
Sample_id,probe_barcode_ids
Sample1,A-A01|A-A02
Sample2,A-B01|A-B02
...