Don’t Fear the REAP-Seq: Simultaneous Measurement of Proteins and RNAs at the Single-Cell Level
Improving cell characterization with single-cell technology
Canonical markers for different cell types have historically been selected for by using antibodies conjugated to fluorophores. This has constrained the amount of proteins that could be measured due to the inherent spectral overlap that is encountered in flow cytometry. Merck and 10x Genomics shattered this limitation with the development of the REAP-Seq assay (RNA expression and protein sequencing). In a recent publication in Nature Biotechnology Peterson et al. used this assay to quantify proteins with 82 barcoded antibodies and more than 20,000 genes in a single workflow, profoundly increasing the level of actionable information acquired compared to previous methods.
How REAP-Seq works
For REAP-Seq, cells are labeled with antibodies conjugated to DNA barcodes (AbBs) instead of with traditional antibodies conjugated to fluorophores. These barcodes consist of 65-66 bp of DNA divided into a NGS Read (used as a primer for amplification and sequencing), a unique 8 bp antibody barcode, and a 24-25 bp poly(dA) sequence. To learn more about how to conjugate antibodies to DNA barcodes, see the Methods section of the publication. These AbBs labeled cells are then subjected to single-cell RNA-seq (scRNA-seq) using the Chromium Single Cell 3’ Solution from 10x Genomics. Paired-end sequencing reads for mRNA and protein are generated and protein sequences aligned using an antibody dictionary that associates each antibody to its unique 8 bp barcode. Then, cells are grouped by their Chromium-generated cell barcodes, resulting in a digital protein and gene expression matrix.
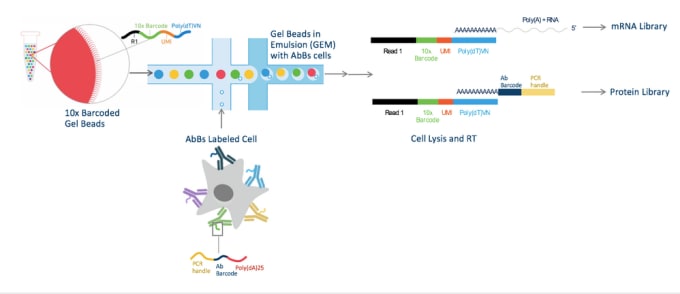
Cells are labeled with antibody barcodes (AbBs) and loaded into the Chromium System. 10x Barcoded Gel beads combine with AbBs cells to form GEMs (Gel Beads in Emulsion). Each functional GEM therefore contains a single AbBs cell, a single Gel Bead, and RT reagents. Within each GEM reaction vesicle, a single cell is lysed, the Gel Bead is dissolved to free the identically barcoded RT oligonucleotides into solution, and reverse transcription of polyadenylated mRNA occurs. At the same time, the AbB sequence can hybridize to the 10x Barcode sequence and be extended to produce shorter protein library cDNAs.
Measuring the immune response in preclinical models
The potential applications of the REAP-Seq assay are abundant, including measuring intracellular signaling pathways or, in combination with perturbation types such as CRISPR, to dissect the mechanisms of different cellular phenotypes. In the publication, the authors demonstrated the utility of REAP-Seq to monitor immune responses in preclinical models by characterizing the in vitro activation of naive CD8+ T cells from donor blood.
The treated (activated) and untreated (control) cells from 3 donors were labeled with a panel of 80 AbBs, live-cell sorted, and subjected to scRNA-seq. For each donor, the treated and untreated samples were merged into a digital gene or protein expression matrix, with unsupervised clustering performed with Seurat. The authors were able to identify genes and proteins differentially expressed in these treated and untreated naive donor CD8+ T cells. The upregulation of a cell proliferation marker in treated cells was consistent with previous findings, however; the unbiased scRNA-seq approach enabled identification of unexpected genes that were also differentially expressed. Protein analysis identified 16 differentially expressed proteins overlapping across the 3 donor samples compared to untreated cells. Notably, a loss of a naive T-cell marker expression and a gain-of-effector memory T cell marker expression was observed, with a substantial number of treated cells expressing CD4 and CD25 (markers associated with regulatory T cells), and confirmed via flow cytometry. Interestingly, mRNA and protein levels did not always correlate.
Importantly, REAP-Seq was used to identify and characterize a small outlier population of cells present within the enriched naive CD8+ T cells. In all donors, there were 17 shared differentially expressed proteins between the outlier cluster and the rest of the cells. When viewing RNA expression, 56 overlapping genes were differentially expressed in this outlier population compared to the rest of the cells, with 3 of the genes also differentially expressed at the protein level. Combining the protein and RNA data revealed that these cells were enriched with genes involved in the transcriptional regulation of megakaryopoiesis, the process whereby mature megakaryocytes develop from the common myeloid progenitor blood cell lineage.
REAP-Seq demonstrates the flexibility of the Chromium Single Cell 3’ System for assay development
By combining protein expression with the unbiased approach of scRNA-seq, the authors were not only able to measure the effects of immune stimulation, but were also able to identify and characterize a previously unknown cell type present in donor cells. The potential uses of the REAP-Seq assay are numerous and demonstrate the flexibility of the Chromium System for multiple single-cell gene expression applications.
Read more about the development of REAP-seq with lead author Vanessa Peterson in our Researcher Spotlight.
Additional Resources
- Read the REAP-Seq paper by Peterson et al. here.
- Learn more about the Chromium™ Single Cell 3' Solution.
- Read Chromium™ Single Cell 3' Solution application notes.
- Check out our data analysis and visualization tools - Cell Ranger™ and Loupe™ Cell Browser.
- Download Chromium™ Single Cell 3' Solution datasets.
