Single cell immune profiling fuels biomarker discovery in clinical cancer samples
*Impact at a glance: What can you discover with both whole transcriptome gene expression data and paired, full-length V(D)J transcript data from single cells? Cancer researchers are using the Universal 5’ Gene Expression assay (previously known as Chromium Single Cell Immune Profiling) to more deeply profile clinical samples and unlock insights that can improve T-cell-based therapies.*
Variability in drug response and side effects between patients remain a major hurdle for cancer drug development—oncology drugs only have a 3–5% success rate in clinical trials (1). Improved strategies for pre-clinical research aimed at identifying biomarkers and drug targets could help boost this sobering number.
Researchers often use flow cytometry and mass cytometry to explore cell diversity, especially in immuno-oncology, but these methods require the use of well-characterized protein markers, limiting the potential for novel biomarker discovery.
Bulk RNA-sequencing (RNA-seq) opened the door to characterizing cells using the whole transcriptome, but it averages gene expression levels across cells, masking the cell-type-specific patterns of gene expression within a tumor (2). Single cell RNA-seq (scRNA-seq) provides a more complete picture, allowing researchers to truly visualize the heterogeneous portrait of tumors.
But even scRNA-seq alone fails to capture a full picture of patient to patient variability. The molecule crosstalk between the tumor and immune cells plays a key role in shaping tumor growth and drug efficacy and toxicity.
Chromium Single Cell Immune profiling allows simultaneous analysis of T-cell receptor (TCR) sequences and gene expression profiles from the same individual cells, enabling researchers to measure clonal expansion and identify often overlooked immune cells in the tumor microenvironment. Improved analysis of the interactions between tumor and immune cells can not only improve predictive and prognostic biomarker discovery, but advance the use of therapeutics into new patient populations.
In a recent Research Snapshot, we highlighted a research study that used our tools to unravel the anti-PD1 response in patient tumor samples of the atypical immunogenic tumor clear cell renal cell carcinoma (ccRCC)—the most common form of kidney cancer.
But one example isn’t enough to convey the capacity of Chromium Single Cell Immune Profiling to enable impactful insights into cancer biomarker discovery. In this blog, we will highlight three additional studies that use our technology to:
- Standardize single cell analysis of frozen patient samples
- Increase the efficacy of tumor-infiltrating lymphocyte (TIL) therapy
- Expand treatment options for acute myeloid leukemia
The nucleus may hold the key to clinical insights in cancer
Single cell immune profiling transformed our understanding of cancer heterogeneity. But using single cell immune profiling to analyze samples during clinical trials presents several challenges.
These analyses typically require large, fresh tissue samples. Not only are patient samples often small, clinicians need to process tissue immediately for profiling downstream, which makes standardized multicenter studies nearly impossible. Due to these limitations, most researchers analyze small patient cohorts, making it challenging to draw statistically significant conclusions that apply to a broad set of patients.
Nuclei isolation bypasses the need for fresh tissue samples. But single nuclei RNA sequencing (snRNA-seq) limits the information researchers can garner from single cell analyses. Using whole cells as input, researchers can analyze both whole transcriptome gene expression data and paired, full-length V(D)J transcript data—transcripts for T-cell and B-cell receptors (BCRs). TCR and BCR transcripts are usually expressed at low levels, even in immune cells. Nuclei isolation excludes RNA in the cytoplasm, so fewer transcripts are captured and analyzed. This issue is amplified in a clinical setting, where tissue samples are sparse.
Researchers from Columbia University recently reported in Nature Genetics that they captured the whole transcriptome and paired full-length TCR transcripts from snRNA-seq analysis of small, frozen tissue samples—such as core needle biopsies—routinely collected during clinical trials (3).
The authors compared Chromium Single Cell Gene Expression 3’ and 5’ analysis of matched fresh and frozen tissues from patients with a variety of cancers, including non-small cell lung cancer (NSCLC), metastatic cutaneous melanoma, and uveal melanoma. Analysis with snRNA-seq using either 3’ or 5’ chemistry of frozen tissue was comparable to that of tissue-matched scRNA-seq of fresh tissue. In some cases, snRNA-seq analysis of frozen tissue using 5’ chemistry outperformed scRNA-seq analysis of fresh tissue using 5’ chemistry. scRNA-seq only recovered 23% of cancer cells in NSCLC samples while snRNA-seq recovered up to 93%.
They then tested how well snRNA-seq using 5’ chemistry could evaluate the cellular makeup of a previously characterized sample type. They analyzed ten-year-old frozen biopsy samples from a participant in the first clinical trial (KEYNOTE-001) using the anti-PD1 antibody MK-3475, also known as pembrolizumab, before and after treatment. The cell populations they found based on known gene expression signatures matched previous single cell analyses of tumor samples from patients with a similar response. They identified changing transcriptional profiles over time, and found a population of cancer cells with a transcriptional signature indicative of immunotherapy resistance enriched in the post-treatment samples.
They also saw increased immune infiltration in the tumor post-treatment with a variety of T cells, including stem-like, precursor-exhausted, and terminally differentiated clonotypes. The researchers also analyzed 169,000 cells from 20 core needle biopsies from seven participants in a multicenter clinical trial testing the efficacy of the MEK (mitogen-activated protein kinase kinase enzyme) inhibitor, selumetinib. They recovered diverse cell populations with expected transcriptional signatures.
The Columbia University scientists are optimistic their snRNA-seq/TCR-seq method will help other clinical researchers interested in addressing questions about immuno-oncology.
“The ability to work from frozen samples opens the door to multi-institutional collaborations that will propel the discovery of biomarkers and drug targets. It also gives us the opportunity to apply cutting-edge computational techniques in the analysis and integration of the unlocked clinical data,” said Elham Azizi, PhD, Assistant Professor of Biomedical Engineering and Herbert and Florence Irving Assistant Professor of Cancer Data Research at the Irving Institute for Cancer Dynamics, who co-supervised this study with Benjamin Izar, PhD, Assistant Professor of Medicine at the Herbert Irving Comprehensive Cancer Center (4).
Izar added, “And because our method requires only a minute amount of tissue, the remainder of the sample may be used for additional studies. This really is a win-win for researchers, clinicians, and, most importantly, our patients.”
Turning tumor cells against themselves
Immune checkpoint inhibitors, such as the anti-PD1 drug nivolumab, block the signals from the immune system and tumors that inhibit TIL therapies. Some researchers isolate TILs directly from patient tumors and put them back in the patient to amplify their tumor-specific immune response. Though TIL therapy is still in its infancy—patients can only access the therapy in clinical trials—it has shown promising effects in people with some cancers, including melanoma, ovarian cancer, and NSCLC (5).
Commenting on the utility of TILs for therapy, Jason Bock, PhD, Vice President of Therapeutics Discovery and Head of Biologics Development at MD Anderson who was not involved in this study, said: “Because TILs come directly from the tumor, they already recognize many targets on the cancer cells. This makes them a very attractive therapy because we don’t have to do anything to point them toward the tumor.”
“That’s different from CAR T cells, for example, which must be genetically engineered to recognize one, or maybe two, targets. A group of TILs taken from a patient’s tumor may recognize many unique targets. This offers a real therapeutic advantage because it prevents the tumor from evading our efforts by hiding one target at a time,” Bock added.
But TILs often aren’t fully active and are eliminated long before they’ve left a lasting impact on the tumor. Researchers are working to modify and expand TILs before they put them back in patients by increasing their activity, amplifying their diversity, and upping their numbers.
Last year, researchers detailed a unique strategy to increase the efficacy of TIL therapy in Molecular Therapy (5). They infected patient tumor cells with a herpes simplex virus 1 (HSV1)-based oncolytic-virus-encoding effector T-cell regulator OX40L and cytokine IL12 to transform the cells into antigen-presenting cells (APCs)—whose primary function is to activate T cells. When they treated patient-derived xenograft and syngeneic mouse tumor models with both the oncolytic virus and TILs, the tumor regressed.
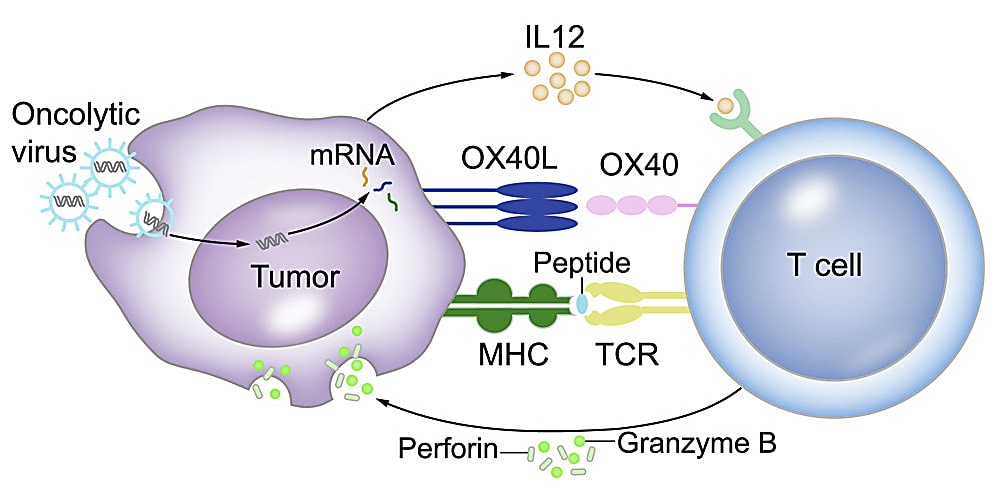
The authors infected primary oral cancer cells isolated from four patients with oncolytic virus and validated their transformation into APCs by co-culture with TILs isolated from the corresponding patient. They confirmed that the tumor cells expressed T-cell binding receptors normally found on APCs, including CD80 and CD86, using quantitative real-time PCR and flow cytometry.
They also used the Chromium Single Cell Immune Profiling kit to conduct scRNA-seq and scTCR-seq and capture a full picture of the TCR repertoire and transcriptome of TILs cocultured with mock- or oncolytic-virus-infected tumor cells. After molecular deconvolution of TCR clonotypes and their corresponding activation status, they confirmed that tumor cells infected with oncolytic viruses contained more activated TILs. Notably, IFN-γ-positive T cells or active T cells were two- to three-fold higher in oncolytic-virus-infected tumor cells than mock-virus-infected tumor cells.
This new oncolytic-virus/TIL combination therapy is a promising candidate for a future clinical trial, especially considering the growing excitement around cancer vaccines. In fact, the backbone of the oncolytic virus used in this study contains the same backbone as T-Vec and teserpaturev, which have been approved for the treatment of melanoma and glioma, respectively (5).
Targeting tumors with a patient’s own T cells
Allogeneic stem cell therapy is currently the only curative option for people with acute myeloid leukemia (AML). But finding a perfectly matched donor is challenging. Some patients have comorbidities that render them ineligible, leading researchers to seek new therapies.
Turning a patient's own T cells against AML bypasses many issues that allogeneic stem cell therapy presents, but blockbuster drugs like PD-1 inhibitors that eradicate solid tumors in many patients haven’t had the effect on AML patients researchers hoped for.
Researchers from MD Anderson wanted to know why. They used Chromium Single Cell Immune Profiling technology to identify biomarkers of response or resistance in bone marrow cells from patients with AML who responded then relapsed or did not respond following treatment with azacitidine and nivolumab—a type of immune checkpoint blockade (ICB) therapy—in a clinical trial (8).
They analyzed 22 bone marrow aspirates from eight participants collected at various time points during their treatment. They performed scTCR profiling on 26,095 T cells from the aspirates before and after ICB therapy and 4,742 T cells from bone marrow aspirates from healthy people. All samples collected from participants had higher TCR clonality than healthy samples.
In the four participants whose disease remained stable or improved after treatment, T-cell repertoires expanded, but retracted in three samples from nonresponders.
The researchers dug deeper to identify specific clonal differences between responders and nonresponders. They performed cluster analysis to identify clonal T-cell populations based on well-characterized gene markers. Notably, they identified a population of CD8+ cytotoxic T cells that expressed Granzyme K (GZMK)—a protein implicated in adaptive immune regulation and apoptosis. This T-cell population was more abundant in the three responders prior to treatment compared to nonresponders pre-treatment.
Using gene set variation analysis for genes associated with T-cell cytotoxicity and exhaustion, the researchers discovered that both exhaustion and cytotoxicity scores of CD8+ cells increased following treatment in responders, but decreased in nonresponders. They discovered that AML with higher expression of GZMK had improved overall survival, suggesting it could be a useful biomarker for ICB therapeutic efficacy in AML patients.
We’re ecstatic that our Chromium Single Cell Immune Profiling technology is helping researchers uncover clinically relevant insights into cancer drug resistance. And this is just the beginning. Learn how to get started with our training videos.
References:
- Wong CH, et al. Estimation of clinical trial success rates and related parameters. Biostatistics 20:273–286 (2019). doi: 10.1093/biostatistics/kxx069
- Yamawaki, TM. et al. Systematic comparison of high-throughput single-cell RNA-seq methods for immune cell profiling. BMC Genomics 22, 66 (2021). doi: 10.1186/s12864-020-07358-4
- Wang Y, et al. Multimodal single-cell and whole-genome sequencing of small, frozen clinical specimens. Nat Genet 55: 19–25 (2023). doi: 10.1038/s41588-022-01268-9
- https://www.eurekalert.org/news-releases/977061
- Ye K, et al. An armed oncolytic virus enhances the efficacy of tumor-infiltrating lymphocyte therapy by converting tumors to artificial antigen-presenting cells in situ. Mol Ther 30: 3658–3676 (2022). doi: 10.1016/j.ymthe.2022.06.010
- https://www.mdanderson.org/cancerwise/what-is-tumor-infiltrating-lymphocyte-til-therapy--6-things-to-know.h00-159460056.html
- Daver N, et al. T-cell-based immunotherapy of acute myeloid leukemia: current concepts and future developments. Leukemia 35: 1843–1863 (2021). doi: 10.1038/s41375-021-01253-x
- Abbas HA, et al. Single cell T cell landscape and T cell receptor repertoire profiling of AML in context of PD-1 blockade therapy. Nat Comms 12: 6071 (2021). doi: 10.1038/s41467-021-26282-z
