Why I chose spatial: Degenerative encephalopathy researcher shares his experience with spatial biology
In Part 1 of the “Why choose spatial” series, we shared insights from our tissue and spatial biology experts regarding key considerations before choosing a solution. In Part 2, researchers share their stories of why they adopted spatial tools into their workflows and what spatial analysis has done to advance their research.
For researchers, adopting a new technology is exciting. We get to learn new skills and, in some cases, play with new gadgets. Even when we’ve done our due diligence in researching the technology, though, applying the method is different than understanding the theory behind it. Therefore, one of the most valuable steps in the evaluation process is connecting with other researchers who have already adopted the technology and learning about their experiences.
We recently spoke with Chenglin Miao, PhD, of Peking University about his adoption of spatial gene expression to investigate degenerative encephalopathy. We hope that sharing his story and insights can help inform your decision as you consider choosing spatial biology for your own research.

Could you please describe your current research projects? How did you become interested in studying your field?
I study the molecular differences in the brain's spatial representation system (hippocampus and entorhinal cortex), with a focus on how aging impacts this system. Specifically, my lab studies place cells in the hippocampus and grid cells in medial entorhinal cortex, which form the basis for quantitative spatiotemporal representation of places, routes, and associated experiences during behavior and in memory.
According to the World Health Organization, the number of people aged over 60 is set to rise to 2 billion by 2050. Although older individuals report substantial declines in their navigational capabilities, most aging research has focused on changes in memory and attention rather than spatial navigation. We know, however, that key structures of the brain’s navigation system are particularly vulnerable to the deleterious consequences of aging. While recording place cells in young and aged mice, we found that aging induced differences in the field sizes, firing rates, stability, decoding accuracy, and other indexes of these cells. Consequently, we became interested in understanding the cellular components of the basic navigational system during the aging process.
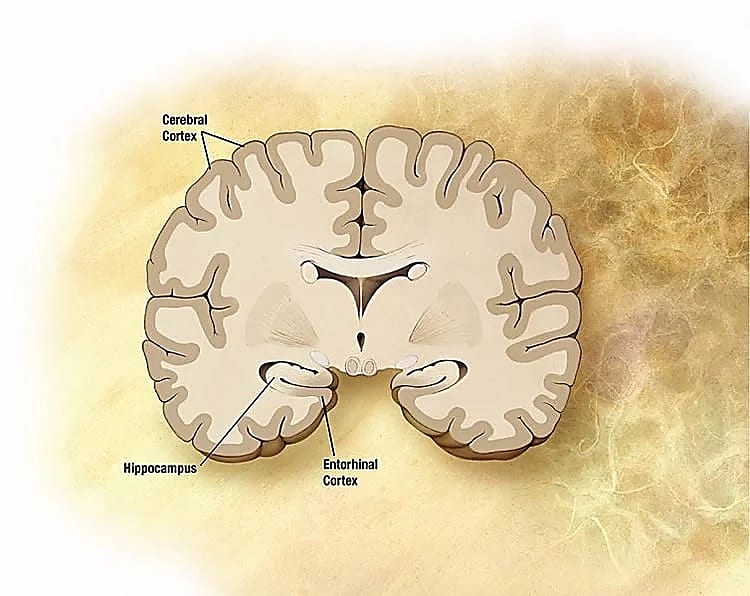
What led you to consider using spatial technology to answer your research questions?
In our mouse model, we want to see synchronous or correlated changes in the entorhinal–hippocampal network that result from aging. Since compartmentation is a basic characteristic of the brain, understanding how different parts work together [in the context of tissues] is crucial.
Were there any technology limitations or experimental challenges that you thought a spatial approach could resolve? What led you to choose Visium over other spatial technologies?
Single cell RNA-sequencing (scRNA-seq) requires the generation of single cell suspensions, which is difficult to produce in aged mice. Visium Spatial Gene Expression uses tissue sections as the input, thus overcoming this challenge of scRNA-seq.
How has taking a spatial approach informed your next research questions or follow-up experiments?
A spatial approach provides a better and more general view of my data, enabling detailed validation and exploration of my scientific questions. Specifically, we want to take advantage of Visium to explore molecular mechanisms that explain differences in the entorhinal–hippocampal representations between young and aged mice. We are currently generating the data and look forward to publishing it in the future.
Stay tuned for future entries in our “Why choose spatial” series as we hear from more researchers about why spatial was a good fit for their research questions, what experimental hurdles it has helped them overcome, and how they’ve used it to make new discoveries in their field of biology.
