For use with: PN-1000481 Xenium Analyzer, 12-Month Warranty
Xenium Onboard Analysis v4.0.1
Patch bugs identified in XOA v4.0.0 and fixed in v4.0.1:
- Fixed rare image deconvolution bug that can cause tiles in the morphology image to be completely black (pe = 0).
- Fixed transcript density heatmap misalignment bug in the Region Details plot of the analysis summary when transcripts are not present in the right side or bottom of the ROI.
Release highlights
Enables the instrument to run Xenium Gene & Protein Expression assay, including:
- Added an interface to select protein subpanels.
- Added XOA v4.0 with support for protein quantification and secondary analysis.
- Improvements to cell segmentation image processing and nucleus segmentation.
Xenium Onboard Analysis v4.0
You must update Xenium Explorer to v4.0 to view data generated with XOA v4.0.
- Upgraded
experiment.xeniumversion to v6.0.
Algorithm updates:
- Improved image processing:
- The 2D multi-focus processing algorithm for morphology images uses the same global focus map for consistent segmentation and quantification.
- Morphology images are deconvolved to remove out-of-focus light and visualize finer details.
- Improved nucleus segmentation: To enable better segmentation in nuclei-dense regions, the algorithm performs joint inference on the 2D DAPI and 18S interior stain images and then adds nuclei segmented from the 3D DAPI Z-stack.
- XOA v4.0 includes image processing, protein quantification, and secondary analysis algorithms to generate Xenium In Situ Gene and Protein Expression with Cell Segmentation Staining assay data (read here). For protein images only, background subtraction accounts for fluctuations in tissue autofluorescence by utilizing two cycles of autofluorescence imaging.
- Secondary analysis of RNA and protein data: Cells with fewer than five transcripts are removed prior to secondary analysis.
Output updates:
- 2D morphology images: The
morphology_focus/file names have been renamed frommorphology_focus_[number].ome.tiftoch[number]_[marker-name].ome.tif. The image metadata contains the offset value that was added to preserve negative values after background subtraction and enable accurate per-cell protein quantification. - The
aux_outputs/directory includes saturation QC masks for all assays in themorphology_focus_qc_masks/directory. - Xenium In Situ Gene and Protein Expression with Cell Segmentation Staining assay datasets contains the same files as a normal XOA output bundle, in addition to protein data-specific output files detailed here.
- For Xenium Protein datasets, the cell-feature matrix has a Protein Expression feature type containing the scaled mean fluorescence intensity. The scaling factor is saved in the matrix h5 file.
- Analysis summary: The Image QC tab gallery headers have been reorganized and renamed. The "Decoding image QC" has been renamed to "RNA Decoding Images". The "Morphology Background QC Gallery" has been renamed to "Autofluorescence Images". The "Morphology Image QC Gallery" has been renamed to "Cell Segmentation Images". The order of images now matches how they are acquired on-instrument and are labeled by cycle number.
- Added new metadata and metrics to the
experiment.xeniumfile.
Xenium Onboard Analysis v3.3
- No changes since XOA v3.3.
Instrument software and user interface
- Enabled instrument to run Xenium Gene & Protein Expression assay.
- Added interface to select protein subpanels for Xenium Gene & Protein Expression assay.
- Updated Load and Unload consumables screens to indicate when Universal Decoding Module A reagents are compatible or required.
- Updated XOA pipeline selection options for Xenium v1 and Xenium Prime assays to enable selection of XOA v3.3 or XOA v4.0.
- XOA pipeline version v2.0 will no longer be supported on XA v4.0+.
- Fixed a rare illumination issue that could cause mid-run failures.
- Incorporated security fixes in instrument firmware.
Xenium Onboard Analysis
- No changes since XOA v3.3.
Instrument software and user interface
- Fixed a bug that resulted in failures during software upgrade.
Xenium Onboard Analysis
- No changes since XOA v3.3.
Instrument software and user interface
- Improved memory usage on instrument to reduce mid-run interruptions.
- Fixed a bug preventing Network Settings page from functioning on some instruments.
- Incorporated security fixes in instrument firmware.
- Updated the Ubuntu kernel to v5.15.0.134.
Xenium Onboard Analysis v3.3.0
- Multimodal cell segmentation algorithm: improved filtering of incorrect boundary-stain segmentations and merging segmentations of boundary-stain images.
- Bug fixes:
- Fixed misaligned Region Details plot in the analysis summary.
- Improved responsiveness of analysis summary.
- Fixed bugs in multimodal cell segmentation to prevent crashes in edge cases. This fix does not impact cell segmentation results.
Instrument software and user interface
- Selection of preservation method is now a required field with cassette details input; dropdown menu has been expanded to include additional options (cells or cell suspension, fixed frozen, and other). This update is intended to more accurately capture sample metadata in outputs such as the analysis summary, and does not currently impact instrument operations or analysis.
- Updated instrument UI to support configuring Network Settings.
- Updated instrument UI to support selecting a local timezone.
- Improved instrument setup steps including timezone selection, network configuration, and local network shared folder setup.
- Improved algorithm for calculating the updated runtime estimation that appears mid-run.
- Fixed bugs to reduce mid-run interruptions and Readiness Test failures.
- Fixed bug impacting DAPI and autofluorescence channel controls during region selection.
Instrument software and user interface
- Fixed bug that in rare instances could cause XOA pipeline to crash for samples with large ROI and high transcript densities.
Xenium Onboard Analysis v3.2.0
-
Algorithm updates for transcript decoding (Does not require batch correction. Learn more here):
- Fixed a Q-Score calibration bug where the lowest raw Q-Score bin only contained transcripts with the minimum raw Q-Score. When this bin did not contain any negative control codewords, the transcripts were calibrated to Q40. In internal data, this affected no more than 1 transcript per dataset.
- Improved puncta signal trace filtering so two real transcripts in close proximity are more likely to be detected.
-
Output format changes:
- Upgraded
experiment.xeniumversion to v5.2. - Added information in
analysis_summary.htmlandmetrics_summary.csvfor Xenium Prime 5K custom panel designs that include boosted pre-designed genes. The output changes are described on this page. - Added a
/density/codewordsmatrix fortranscripts.zarr.zipto enable visualization of gene splitting for Xenium Prime 5K data in Xenium Explorer v3.2.
- Upgraded
-
Bug fixes:
- Fixed bug in
analysis.zarr.zipcluster indices that caused Xenium Explorer to display incorrect cell count for final graph-based cluster.
- Fixed bug in
Instrument software and user interface
- Improved runtime estimation, decreasing the chance of underestimate.
- Updated instrument run progress display after estimated time has elapsed to clarify run status.
- Updated Instrument UI to show estimated time of completion for overview scan.
- Updated Instrument UI to show run chemistry (v1 or Xenium Prime) on the run summary page and imaging configuration screens.
- Fixed rare bug leading to increased runtime for large samples.
- Fixed bug leading to appearance of Ubuntu upgrade pop-up.
Xenium Onboard Analysis v3.1.0
- Output format changes:
- The
cell_feature_matrix.zarr.zipis zstd compressed and includes a compressed sparse column (CSC) matrix representation for enhanced performance of certain features in future Xenium Explorer releases. - Upgraded
experiment.xeniumversion to v5.1 with new fields:panel_designer_version,num_transcripts,num_transcripts_high_quality, andnon_zero_matrix_entries analysis_summary.htmlPanel Specifications table includes the "Panel designer version" specifying the version of Xenium Panel Designer software used to generate a custom panel.metrics_summary.csvvalues truncated to 15 decimal places.
- The
- Bug fix:
- Fixed a TIFF compression artifact related to over-quantization in tiles with a high number of zero-value pixels. The artifact is usually seen in areas without tissue.
- Fixed a bug that caused background QC images in the analysis summary to have slightly higher intensities than were actually used for background subtraction.
- Fixed a rare metric calculation bug that could cause the pipeline to crash.
Xenium Onboard Analysis v2.0.2
- Bug fix: Fixed a TIFF compression artifact related to over-quantization in tiles with a high number of zero-value pixels. The artifact is usually seen in areas without tissue.
Instrument software and user interface
- Fixed a critical bug resulting in a rare image handling error.
- Updated Instrument UI to support export of run data to network shared folders.
- Updated Instrument UI to enable setup of network shared folders.
- Updated Instrument UI to enable direct export of run data to a USB drive.
- Improved Instrument UI for importing custom panel JSON files.
Instrument software and user interface
- Fixed an intermittent bug with touchscreen flashing and carrier getting unlocked when door is open.
Instrument software and user interface
- Fixed a bug intermittently showing an error message after updating to v3.0 software.
- Fixed a bug with uploading custom panels via the Menu Settings.
Security updates
- Addressed CVE-2024-6387.
Xenium Prime analysis requires XOA v3.0. For Xenium v1, we recommend using XOA v3.0 software. However, if you are in the middle of an ongoing Xenium v1 study with the XOA v2.0 software, v2.0.1 is available to select.
Xenium Onboard Analysis v3.0
You must update Xenium Explorer to v3.0 to view data generated with XOA v3.0.
- Upgraded
experiment.xeniumversion to v5.0.
Algorithm updates for image processing and transcript decoding
- Improvements to the Image Processing Algorithm for RNA fluorescence images. This improvement leads to an increase in the maximum decodable transcript density for both Xenium v1 and Xenium Prime assays. See Image Processing Algorithms page for more details.
- Genomic control metrics are output when the gene panel includes genomic control probes. The genomic control probes were added to Xenium Prime panels for improved false discovery rate estimation.
- (Note added 9/19/24) Cell segmentation: Improved multimodal cell segmentation by filtering incorrect segmentations of the boundary-stain images.
Output file changes
- The
cells.csv.gzandcells.parquethave new columns:segmentation_method,genomic_control_counts,nucleus_counts. - The
transcripts.csv.gzhas been removed from XOA outputs. Use thetranscripts.parquetfile for faster loading and optimized performance on large datasets. - The
transcripts.parquethas new columns for simple filtering of transcripts:is_gene,codeword_category. - In order to reduce the file size of
transcripts.parquet, the xyz location coordinates are now quantized into discrete values,nucleus_distanceis in 15 nm steps, andqvscores are rounded down to the nearest 0.25. In addition, the file is zstd compressed. - Updated some metrics in the
analysis_summary.htmlandmetrics_summary.csvto enhance QC experience:- Changed calculation for the transcript density metric
decoded_transcripts_per_100um2to only use transcripts in cells (labeled as "Cellular transcripts per 100 µm2" in the analysis summary). - For Xenium Prime data, the following genomic DNA metrics were added: "Adjusted genomic control probe rate", "Genomic control probe counts per control per cell", "Estimated number of false positive transcripts per cell including genomic counts". They are calculated using the genomic control probes. For more details, see the Analysis Summary Overview.
- Changed calculation for the transcript density metric
- Updates to
analysis_summary.htmlplots:- On the Summary tab, the Region Details section includes density plots for Transcript density map, Mean Q-score, and Negative controls. See Analysis Summary Overview for more information on plot usage and interpretation.
- On the Decoding tab, improved Counts per Gene plot and added Gene Count table to accommodate panels with more genes. Includes ability to search for specific genes and sort by name or transcript count. The Transcript Quality plot has been removed. The Mean Q-score Spatial Map is now on the Summary tab in Region details.
- On the Analysis tab, removed the UMAP dropdown option to display K-means clusters. The K-means clustering results are still available in the secondary analysis outputs.
- On the Image QC tab, added a gallery of morphology background images that are subtracted from the raw stain images during the multi-stain image acquisition process. The background images are also available in the
aux_outputs/background_qc_imagesdirectory.
- To optimize performance, the
cell_feature_matrix.zarr.zipis written out in chunks. Thetranscripts.zarr.zipchanged data stored ingrids/array to enable the new scaled transcript view in Xenium Explorer v3.0. - Added new metadata and metrics to the
experiment.xeniumfile. - Tiny XOA v3.0 output bundle examples added to the example datasets page to help test code dependencies. These examples are trimmed down to a few FOVs from larger datasets.
Bug fixes
- Fixed a bug that could cause the pipeline to crash in extremely rare situations.
- Fixed a bug to ensure interior stain-based expansion extends all the way to the neighboring cell’s boundary.
- Fixed a bug to guarantee every nucleus and cell segmentation mask is a single connected component.
Xenium Onboard Analysis v2.0.1
XOA v2.0.1 software includes all the changes noted in XA v2.0 release notes. A few bug fixes are included in the v2.0.1 release. These fixes are not expected to introduce any batch effects for ongoing studies using XOA v2.0 software. The XOA v2.0 documentation is available here.
Bug fixes
- Fixed a bug that could cause the pipeline to crash in extremely rare situations.
- Fixed a bug to ensure interior stain-based expansion extends all the way to the neighboring cell’s boundary.
Instrument software and user interface
- Enabled instrument to run Xenium Prime assays.
- Added option to select whether Xenium Prime reagents are being used on the Run settings screen.
- Added the pre-designed Xenium Prime 5K Human Pan Tissue & Pathways Panel and Xenium Prime 5K Mouse Pan Tissue & Pathways Panel
- Updated Instrument UI to ensure compatible Panels are selected based on assay reagents selected (Xenium Prime or Xenium v1).
- Enabled option to select one of two XOA pipeline versions for Xenium v1 panels (XOA v2.0.1 or XOA v3.0) on the Assay configuration screen.
- Improved runtime estimation.
- Updated UI to display Xenium Software release notes on homepage when a new upgrade is available.
- Deprecated previously unassigned codewords for two Xenium v1 pre-designed panels: Xenium Human Brain Gene Expression Panel (v1.2) and Xenium Human Breast Gene Expression Panel (v1.1).
Xenium Onboard Analysis
- You must update Xenium Explorer to v2.0 to view data generated with XOA v2.0.
- Upgraded
experiment.xeniumversion to v4.0.
- Upgraded
- Output file changes
- The improved algorithm for generating 2D focus images now outputs files in multi-file OME-TIFF format, instead of the
morphology_focus.ome.tifandmorphology_mip.ome.tiffiles. The newmorphology_focus/directory contains the 2D focusmorphology_focus_xxxx.ome.tiffiles. For DAPI-only datasets, the directory containsmorphology_focus/morphology_focus_0000.ome.tif. For cell segmentation staining workflow datasets, there are fourome.tiffiles, one per stain image in this directory. - Updated the
analysis_summary.htmlandmetrics_summary.csvto enhance QC experience- Added segmentation metrics reporting number and percentage of cells segmented by each method in Cell Segmentation tab.
- The Image QC tab includes a morphology image gallery for cell segmentation staining workflow data.
- A new metric, "Nuclear transcripts per 100 µm2", replaces "Decoded transcripts per 100 µm2" in Key Metrics. The nuclear transcript density metric can be more easily compared between datasets analyzed with different cell segmentation algorithms.
- "Fraction empty cells" renamed to "Percentage of empty cells" in the analysis summary.
- Added fields to
experiment.xeniumandanalysis_summary.htmlto include cell segmentation method information. - In
cells.zarr.zip, moved polygon vertices intopolygon_setsgroups, which include acell_indexfield to support cells with zero nuclei or more than one nuclei. - Added
codeword_indexcolumn to transcripts CSV and parquet files. - Added
label_idcolumn to nucleus and cell boundary CSV and parquet files to help distinguish the nuclei in multinucleate cells.
- The improved algorithm for generating 2D focus images now outputs files in multi-file OME-TIFF format, instead of the
- Algorithm updates for image processing and segmentation:
- Default isotropic nuclear expansion distance reduced to 5 µm instead of 15 µm for nuclear expansion-only cell segmentation and multimodal cell segmentation.
- Multi-tissue stain image alignment and background subtraction.
- Multi-focus image fusion algorithm for projecting tissue stain images to 2D (DAPI-only or Multi-Tissue Stain Kit Mix).
- Updated nucleus segmentation model to reduce false positive detection in tissues with bright non-nuclear DAPI signal, such as skin.
- New multimodal cell segmentation methods for Xenium In Situ Gene Expression with Cell Segmentation Staining workflow.
- Tiny XOA output bundle examples are now provided on the example datasets page to help test code dependencies. These examples are trimmed down to a few FOVs from larger datasets.
Instrument software and user interface
- Added an assay configuration page to the interface for selecting cell segmentation protocols, enabling the use of the Cell Segmentation Staining assay and Xenium Cell Segmentation Staining reagents and detection kits for multimodal cell segmentation.
- Added the pre-designed Xenium Human Immuno-Oncology Profiling Panel.
- Reduced exposure times to avoid saturation in crowded regions.
- Remote access for 10x support is automatically disabled after 48 hours.
- Enabled upload of custom panels from the Settings page.
- Improved error reporting and telemetry.
Xenium Onboard Analysis
- No changes since XOA v1.7.
Instrument firmware
- Improved error reporting.
- Improved compute resource usage to reduce run times.
Instrument user interface
- Region selection caps number of ROIs to eight per slide.
Xenium Onboard Analysis
- No changes since XOA v1.7.
Instrument firmware
- Added a USB copy tool to improve process for output data transfer.
- Improved hydration cycle start logic.
- Improved error reporting.
- Added write-only file to improve handling of network credentials.
Instrument user interface
- Updated region selection warning to indicate a risk of run failure when more than eight ROIs are selected.
Xenium Onboard Analysis
- Xenium Explorer v1.2 or later is required to view data generated with XOA v1.7.
- Upgraded
experiment.xeniumversion to v3.0.
- Upgraded
- Output file changes
- Added
aux_outputs/overview_scan_fov_locations.jsonandoverview_scan.pngimage to Xenium output bundle. Renamedaux_outputs/fov_locations.jsonfile toaux_outputs/morphology_fov_locations.json. Auxiliary output files are described here. - The
analysis_summary.htmlfile has a new Image QC tab with a gallery of RNA images from each cycle and channel (same images as the higher-resolution images inaux_outputs/per_cycle_channel_images/). - Gene names renamed from
BLANK_xxxxxtoUnassignedCodeword_xxxxxin transcripts (.zarr.zip,.csv,.parquet) andcell_feature_matrix.h5output files.
- Added
- Bug fixes
- Improvements to reduce the potential for FOV artifacts in areas with extremely high transcript count. Note: For samples prepared following the FFPE and fresh frozen Tissue Preparation Handbooks and used with panels designed under the detection budget for the samples' tissue type, decoding results will not be impacted. For samples exceeding detection budget, this change will impact decoding results by making transcript count more uniform across FOVs. Read more about detection budget on the Getting Started with Xenium Panel Design page and the Xenium Add-on Panel Design Tech Note.
- Updated
metrics_summary.csvto contain the same metrics as theanalysis_summary.html. - Fixed cell segmentation bug where elongated cells with straight edges were sometimes produced at an FOV boundary.
- Further reduced occurrence of global stitching failures due to imaging of empty FOVs.
- Several minor bug fixes in the analysis summary UI.
Instrument firmware
- Improved error reporting.
Instrument user interface
- Includes the pre-designed Xenium Human Colon and Xenium Human Skin Gene Expression panels.
- Pre-designed Xenium Human Lung Gene Expression Panel (v1.1): deprecated three previously unassigned codewords.
- Improved UI for software upgrade download and installation.
- Added a checkbox prompt to check reagent bottles are loaded.
- Improved how the "Number of targets" are displayed on the panel selection interface.
- Added a warning to avoid special characters in region names.
- Improved interface animations and screen transitions.
Xenium Onboard Analysis
- Output file changes
- Added
aux_outputs/per_cycle_channel_images/directory to Xenium output bundle. It contains downsampled RNA images (maximum intensity projection) from each cycle and channel. - Added
panel_predesigned_idtoexperiment.xeniumand upgradedpatch_versionto 2.0.2.
- Added
- Improvements to
metric_summary.csv:- "Panel_name" switched from the panel file name to the panel name specified by the JSON file.
- Added "design_id" and "predesigned_panel_id".
- Added "thickness_transcripts_high_quality".
- Added metrics for Xenium Ranger
import-segmentationpipeline results.
- Improvements to the
analysis_summary.html:- Alert thresholds added to alert help text.
- Removed alert for "Adjusted negative control probe rate" (see this article for more information).
- Summary tab:
- Added hero metric: "Total high quality decoded transcripts".
- Metadata displayed if data was reanalyzed with Xenium Ranger.
- Decoding tab:
- Added "Thickness of high quality decoded transcripts" metric.
- Added negative control metrics: "Negative control probe counts per control per cell" (warning and error alerts at 0.02 and 0.05, respectively) and "Estimated number of false positive transcripts per cell".
- Added "Unassigned Codewords" category to "Transcript Quality" and "Gene Specific Transcript Quality" plots.
- Added "Mean Q-score Spatial Map" plot, which is useful for checking that data does not contain FOV artifacts
- Bug fixes
- Reduced occurrence of global stitching failure due to the presence of empty FOVs.
- Fixed a bug in the image registration algorithm where it may have failed to produce the correct alignment for nearly empty FOVs (i.e., at tissue edges) in XOA v1.5.
Instrument firmware
- Performance improvements resulting in an approximately 10% faster run time (50 hours, 2 slides, 2x1 cm2 tissue area imaged per slide).
- Improved error reporting.
- Fixed run cancellation handling to ensure buffer exchanges are complete and sample stays hydrated.
- Resolved intermittent problem handling homing of syringe pumps.
- Improved reagent plate deck temperature control checks during Readiness Test.
- Improved auto-recovery of the instrument when the internal control computer is not reachable during bootup.
- Improved liquid cooling fan signal checks.
- Improved system memory footprint.
Instrument user interface
- Includes the pre-designed Xenium Mouse Tissue Atlassing Panel.
- Enabled customers to self-serve system upgrades.
Instrument firmware
- Performance improvements resulting in an approximately 55 hour run time (2 slides, 2x1 cm2 tissue area imaged per slide).
- Improved robustness of objective drying motion algorithm.
- Fixed issue to relax stringent tolerance on motion sensor that resulted in a run failure.
- Resolved false-positive pressure sensor error in the Readiness Test.
- Resolved error causing incorrect reporting of temperature control error, leading to Readiness Test failure.
- Resolved intermittent issue where door fails to unlock after cleanup upon door closure.
- Resolved intermittent problem detecting cassette presence.
- Disabled low use reagents temperature control systems after Readiness Test to prevent condensation.
Instrument user interface
- Includes the pre-designed Xenium Human Multi-Tissue and Cancer Panel.
- Pre-designed Xenium Human Brain Gene Expression Panel (v1.1): deprecated seven previously unassigned codewords.
- Pre-designed Xenium Mouse Brain Gene Expression Panel (v1.1): deprecated the codeword for Lyz2.
- Resolved issue where users could not toggle the UI button to enable remote access for support.
- General improvements for UI and user experience.
Xenium Onboard Analysis
- Output file changes
- Upgraded
transcripts.zarr.zipversion to 4.0. Added attributescodeword_identity,codeword_gene_names,codeword_gene_identity,codeword_count. - Support added for the
Deprecated codewordfeature type (added totranscripts.zarr.zip,cell_feature_matrixfiles,cells.csv.gz, andcells.parqet). Deprecated codewords are not used in the Xenium Onboard Analysis pipeline. Deprecated codewords will appear in Xenium Explorer (until next version) asDeprecatedCodeword_<ID>. - Upgraded
experiment.xeniumversion to 2.0.1 and addedanalysis_uuidmetadata field.
- Upgraded
- General improvements
- Algorithm improvements to increase RNA transcript sensitivity 10-20%, and up to 40% in high-transcript density regions.
- Bug fixes and miscellaneous changes
- Improvements to Panel Specification information provided in the
analysis_summary.html. Panel types will be displayed as "Predesigned", "Add-on Custom", or "Standalone Custom". For add-on custom panels, the "Predesigned Panel ID" will be shown along with the add-on panel "Design ID". - Fixed "Math domain error" failure during image stitching in onboard analysis.
- Improvements to Panel Specification information provided in the
Instrument firmware
- Performance improvements resulting in decreased run time by up to 10 hours.
- Sipper motion enhancements: Optimized speed profiles to minimize vibrations and noise caused by resonance.
- Fixed issue where canceling a run immediately after starting would result in a persistent on-screen error preventing cancellation.
- Fixed issue that caused miscalibrations to generate a critical "not-empty" error for an empty objective wetting consumable.
Instrument user interface
- Includes the pre-designed Xenium Human Brain and Xenium Human Lung Gene Expression panels.
- Improved run time estimate.
- Improved interface to prevent the entry of duplicate panel names or panel names that conflict with pre-defined panels.
- Added an additional tab in the Settings menu to provide information on the last updated firmware version and analysis versions.
- Enhanced user feedback for invalid characters in the run name and cassette name fields to address inconsistencies where certain invalid characters were incorrectly allowed, while certain valid characters were incorrectly flagged as invalid. Run name values are now restricted to ASCII characters, excluding the following characters:
!@#$%&*)+=. - During bootup time, the Run button will remain disabled if additional time is needed to ready the camera and the UI will say:
Please wait a few minutes until the "Start Run" button is active. - Fixed intermittent functionality issue with the "Run Details" button in Settings > Runs (tab on the sidebar). Users can now consistently access run details when the button is pressed.
- Fixed issue on the Region Selection Screen where deleted regions that were previously marked as overlapping would still be selected in the new region selection process. Now, only the desired regions are selected, eliminating any incorrect or overlapping selections.
- Fixed an issue where the region overlap on screen pop-up warning message displayed the original/default overlapping region name instead of the renamed one on the Region Selection Screen.
- Fixed issue where deleting all regions would remove the grid overlay in the Region Selection Screen.
- Fixed issue where the customer interface would freeze when a critical error message appeared on the screen.
- Fixed issue where the UI showed the incorrect current time in the run estimate countdown timer while it computed the estimated run time at the start of the run.
- UI no longer allows identical slide ID names, which used to result in output bundle failures.
Xenium Onboard Analysis
- Output file changes
- The names of the negative control metrics were updated to better reflect how they are calculated (now
Adjusted negative control codeword rateandAdjusted negative control probe rate). These metrics are reported in theanalysis_summary.htmlandmetrics_summary.csvfiles. - Improvements to the
analysis_summary.htmlDecoding tab:- The Gene Counts plot was renamed to Counts per Gene.
- The Gene-Specific Transcript Quality plot now shows the mean calibrated quality of gene transcripts on the y-axis instead of median (see note in v1.2 Xenium Onboard Analysis release notes).
- The plot color palettes were updated so that genes/controls are consistent between panel types and are color-blind friendly. Plot legends were updated to be consistent between plots.
- Output bundle completion marker file (
.end-of-run) added to the output bundle folder. It enables automated script actions to initiate possible data transfer.
- The names of the negative control metrics were updated to better reflect how they are calculated (now
- General improvements
- Improvements to make the FOV stitching algorithm more robust:
- Filter out invalid algorithm features at image boundaries.
- Use orientation, scale, and position to improve algorithm feature-matching
- Use image-based metric (correlation) to detect alignment failure.
- Pairwise registration outliers are removed from global alignment metric to reduce false alerts.
- Improvements to make the FOV stitching algorithm more robust:
- Bug fixes and miscellaneous changes
- Fixed issue to resolve run failures when an OME-TIFF file is created during an intermittent internet connection interruption.
- Panel type "Add_on" now displayed as "Add on" in the
analysis_summary.htmlPanel Specification section. - Fixed a bug where the
z_levelattribute was not reported in µm for some cells in thecells.zarr.zipoutput file.
Instrument firmware
- Improved objective wetting consumable (OWC) calibrations during load consumables to account for drifts.
- Improved various power management controller firmware and reset procedures to reduce errors in Readiness Tests.
- Improved Readiness Test to reduce false-positive failures due to bright rooms.
- Fixed issue where OWC always showed 1 of 1 detected during consumables loading, regardless of its presence.
- Fixed issue in assay performance test (APT) and improved diagnostics for monitoring in APT.
- Fixed overview scan estimate time to the current expectation of 50 minutes.
- Various robustness improvements to handle communication errors in motion subsystems.
- Fixed intermittent motion errors during Objective Cleaning.
- Improved checks and resolution of potential camera link failures during boot time.
Instrument user interface
- Fixed issue where gene panel details were not displayed when switching from load consumables page to import panel page.
- Fixed issue where after reboot, the user interface (UI) would display the registration page even if the device was already registered.
- Fixed an issue where the UI would show a connection error following a black UI screen after dismissing the initial connection error.
- Fixed issue where the instrument door would not open when navigating to the "Remove Consumables" screen.
- Updated "Last Updated" text fields to reflect the correct version of software.
- Fixed issue where, after two errors occurred in the UI, an "Error" pop-up would appear stating that the error had been resolved, but the same errors would be displayed again later in the same menu.
- Additional message on the UI screen for added boot time if camera link failure checks and resolution required.
- The UI now shows software upgrade status in the settings screen.
- The UI now warns and prevents users from entering identical ROI name submissions, identical panel names, and identical Slide IDs for both the Left Cassette and Right Cassette.
- The UI now shows warning text and disables Start Run button in Cassette subpage if the user uses a Cassette name or Run Name containing invalid characters or exceeding 33 characters.
Xenium Onboard Analysis
- Xenium Explorer v1.1 is required to open datasets generated with Xenium Onboard Analysis v1.3 due to a new format for cell IDs in output files.
- Upgraded
experiment.xeniumversion to 2.0 (for Xenium Explorer compatibility).
- Upgraded
- Output file changes
- Cell ID changes:
- To prevent mistakes related to ambiguous data types (e.g., integer vs. string) and to maintain compatibility with single cell tools, the cell ID was changed from a cell index integer to a cell prefix and dataset suffix (e.g.,
cmlbdfdf-1), analogous to Cell Ranger cell barcode IDs. The cell prefix is determined from the cell segmentation polygon, so the same segmentation is always assigned the same cell prefix. This format change impacts all output files that havecell_idinformation, and will be visible in Xenium Explorer v1.1 when cells are selected. - In
cells.zarr.zip, the cell index is now calledseg_mask_value(previouslycell_id). This array contains an index for each cell in the segmentation mask images for a given experiment.
- To prevent mistakes related to ambiguous data types (e.g., integer vs. string) and to maintain compatibility with single cell tools, the cell ID was changed from a cell index integer to a cell prefix and dataset suffix (e.g.,
- Added
cell_idto thecell_feature_matrix.zarr.zipfile. - Added
nucleus_distancecolumn totranscripts.csv.gzandtranscripts.parquetfiles.
- Cell ID changes:
- General improvements
- Updated cell segmentation model to work on more diverse tissue types by training on human brain and human lung data.
- Improved the
analysis_summary.htmlinterface and metric/plot/alert descriptions. - Flipped the X-Y axes on the
analysis_summary.htmlGene Counts plot (Decoding tab) to make reading gene names easier.
- Bug fixes and miscellaneous changes
- Removed redundant information in
cells.zarr.zip(top-levelhomogeneous_transformarray,cell_boundary_polygon_set_idxfield).
- Removed redundant information in
Instrument firmware
- Improved handling of instrument door locking and unlocking during consumables loading.
- Enhanced Readiness Tests to ensure the power management controller and other embedded controllers are in a healthy state of operation prior to the start of a run.
- Fixed various air displacement pipette (ADP) tip and instrument controller false-positives during readiness checks.
- Resolved sporadic camera link failures in Readiness Tests.
- Improved imaging quality by trimming the field of view (FOV) size. FOV demarcation (row x columns) changed from 28 x 19 to 33 x 19 FOVs. FOV size changed from 4240 x 2960 pixels to 3520 x 2960 pixels.
- Intelligent carrier load-position calibrations to prevent incorrect triggering of door-open safety mechanism.
- Guidance to users to remove tip during Readiness Tests to prevent false-positives in instrument readiness.
Instrument user interface
- Improved image and instructions in UI to provide better guidance on loading consumables.
- New UI dialog to warn users to lock the front panel before starting the cleanup process.
- Fixed flickering green checkboxes during objective wetting consumable (OWC) loading.
- Fixed add-on panel targets count, which was incorrectly displayed in UI.
- Removed persistent error messages in Readiness Tests once the error is resolved.
Xenium Onboard Analysis
-
Output file changes
- Added field of view (FOV) information (
fov_namecolumn) totranscripts.csv.gzandtranscripts.parquetfiles. FOV names use chessboard nomenclature. - Added
aux_outputs/fov_locations.jsonfile to Xenium output bundle. It contains the global coordinates for FOVs. - Added versioning to
experiment.xeniumfor file compatibility checking in Xenium Explorer.
- Added field of view (FOV) information (
-
General improvements
- Improved transcript decoding algorithm to reduce FOV-to-FOV variation.
- Calculated Q-Scores are more accurate and precise. Based on internal analysis, there is a consistent small drop in per-gene mean Q-Scores (left plot) between v1.1 and v1.2 outputs. This difference is exaggerated in the median calculation of Q-Scores (right plot).
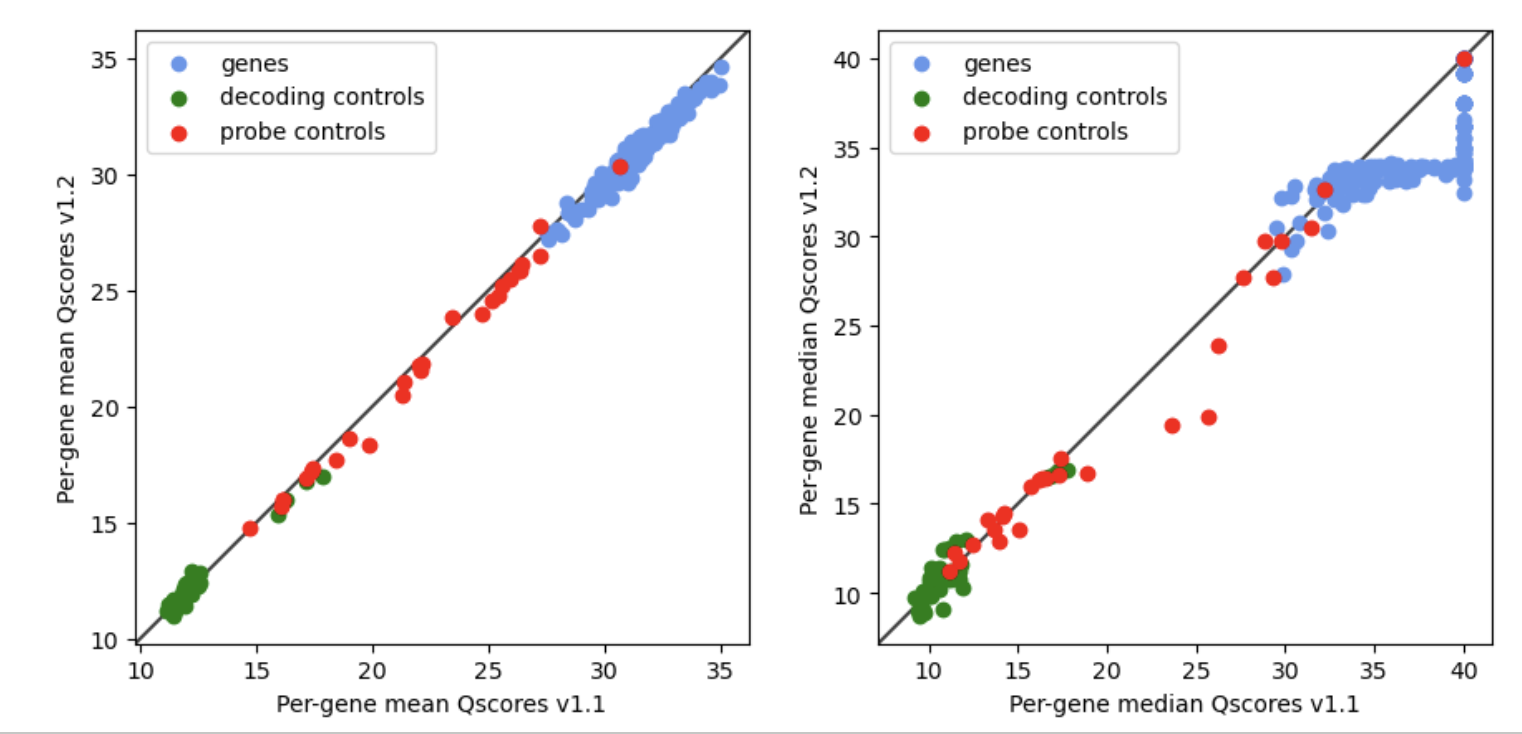
Consequently, the median Q-Scores in the
analysis_summary.htmlGene-Specific Transcript Quality plot may look lower (Decoding tab). This plot will switch to using mean Q-Scores in a future version. -
Bug fixes and miscellaneous changes
- Changed "GEX" in panel names to "Gene Expression".
- Added instrument metadata (
calibration_uuid) toexperiment.xenium.
v1.1 (February 2023)
Instrument firmware
- Users are informed when their samples are no longer being hydrated, ensuring proper care and preservation of their samples.
- Improved Readiness Test procedure to provide additional time for the camera sensor to cool down in high temperature environments.
- Improved calibration procedures.
- Fixed error from attempting to cancel a run while loading consumables when the instrument door is open.
- Improved accuracy in Readiness Tests.
Instrument user interface
- Enabled custom gene panels.
Xenium Onboard Analysis
- Output file changes
- Renamed "Blank Codeword" feature type in cell-feature matrix to "Unassigned Codeword".
- Added
unassigned_codeword_countscolumn tocells.csv.gzandcells.parquetfiles. - Increased chunk size of precomputed densities in
transcripts.zarrfor faster loading in Xenium Explorer.
- General improvements
- Improved run time estimation accuracy.
- Bug fixes and miscellaneous changes
- Fixed
analysis_summary.html(Instrument software versionfield) andexperiment.xenium(instrument_sw_versionfield) to show correct instrument software version. - Based on additional experimental data, thresholds for
high negative control probe ratewarning and error alerts were increased from 2% to 4% and 5% to 10%, respectively, in theanalysis_summary.html. - Various styling improvements to
analysis_summary.html. - Minor bug fixes.
- Fixed