The analysis_summary.html file generated by the Xenium Onboard Analysis pipeline should be among the first output files a researcher looks at after running the Xenium instrument. This file serves three purposes:
- Provides a convenient portable overview of data
- Highlights summary metrics and plots that researchers can use to quickly QC data
- Contains information that 10x Genomics support can use for troubleshooting
Where can I find the analysis summary?
The analysis_summary.html file can be found on the instrument after the run completes (see the Xenium Analyzer User Guide).
- After the run is complete, data generated across all the runs can be accessed under Menu > Open Settings > Runs.
- Click Open Output Folder Location to access the top-level output folder on the desktop. Click the individual runs to open a run-specific screen.
- To access the region-specific output folder, click Open Region Folder.
- The
analysis_summary.htmlis available in the View Analysis Summary folder.
The analysis_summary.html file can be viewed on the instrument, on any web browser, and in Xenium Explorer.
At the top of the analysis_summary.html, there is a banner with the Run name and Region name, which were inputs on the instrument during the Initialize Instrument step (see the Xenium Analyzer User Guide). The Run start time, which is time-stamped by the instrument, is also shown (in UTC). There are five tabs: Summary, Decoding, Cell Segmentation, Analysis, and Image QC.
The default view begins on the Summary tab. When certain metrics fall below key thresholds, warning or error alerts will be shown below these tabs.

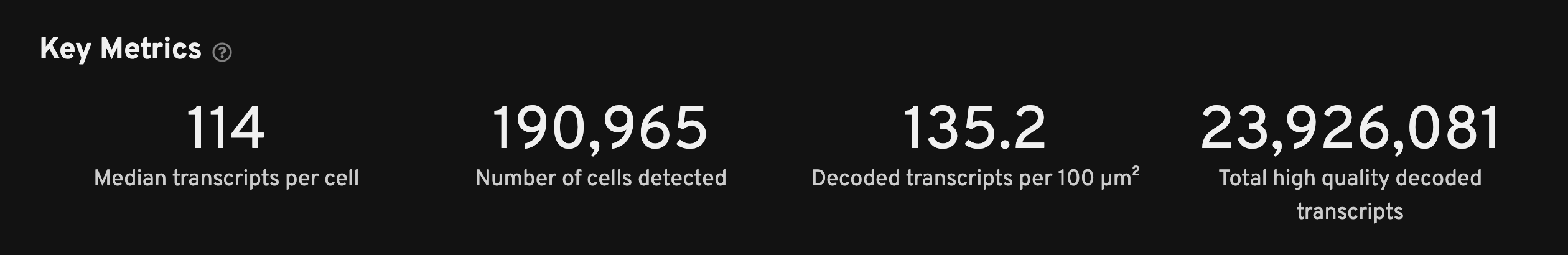
The Key Metrics panel is the starting point to QC your data and to spot outliers among multiple runs at a glance. These metrics are often helpful indicators if there is a problem with the data.
There are no universal thresholds for these metrics as interpretation requires some understanding of the sample and gene panel used. Researchers should have some understanding of how many cells are expected given the tissue size and type. Here are some important considerations for interpreting key metrics:
- Median transcripts per cell and decoded transcript density will depend on tissue type and the selected gene panel (e.g., a 300-gene panel will have more transcripts than a 50-gene panel).
- The number of cells detected will depend on tissue type and the size of the region of interest (ROI) selected on the instrument in the analysis.
| Metric | Description |
|---|---|
| Median transcripts per cell | The median number of transcripts per cell. Cells with zero transcripts are excluded from the calculation. This metric is expected to be lower if the selected gene panel does not match the tissue type. |
| Number of cells detected | The expectation for this metric depends on the tissue sample. |
| Decoded transcripts per 100 µm2 | Counts the number of high-quality decoded transcripts and divides it by the total estimated tissue area to get a decoded transcript density. This metric provides an estimate of transcript density that is independent of cell segmentation, which is performed algorithmically. A warning alert will be shown if there are fewer than 10 transcripts per 100 µm2. |
| Total high quality decoded transcripts | The total number of decoded gene transcripts that decode with high quality (≥ Q20). |
The Sample Region Summary (left) shows the Region name, Slide ID, and Cassette name, which are required inputs during instrument initialization (see the Xenium Analyzer User Guide). Preparation method, also input during instrument initialization, is optional.
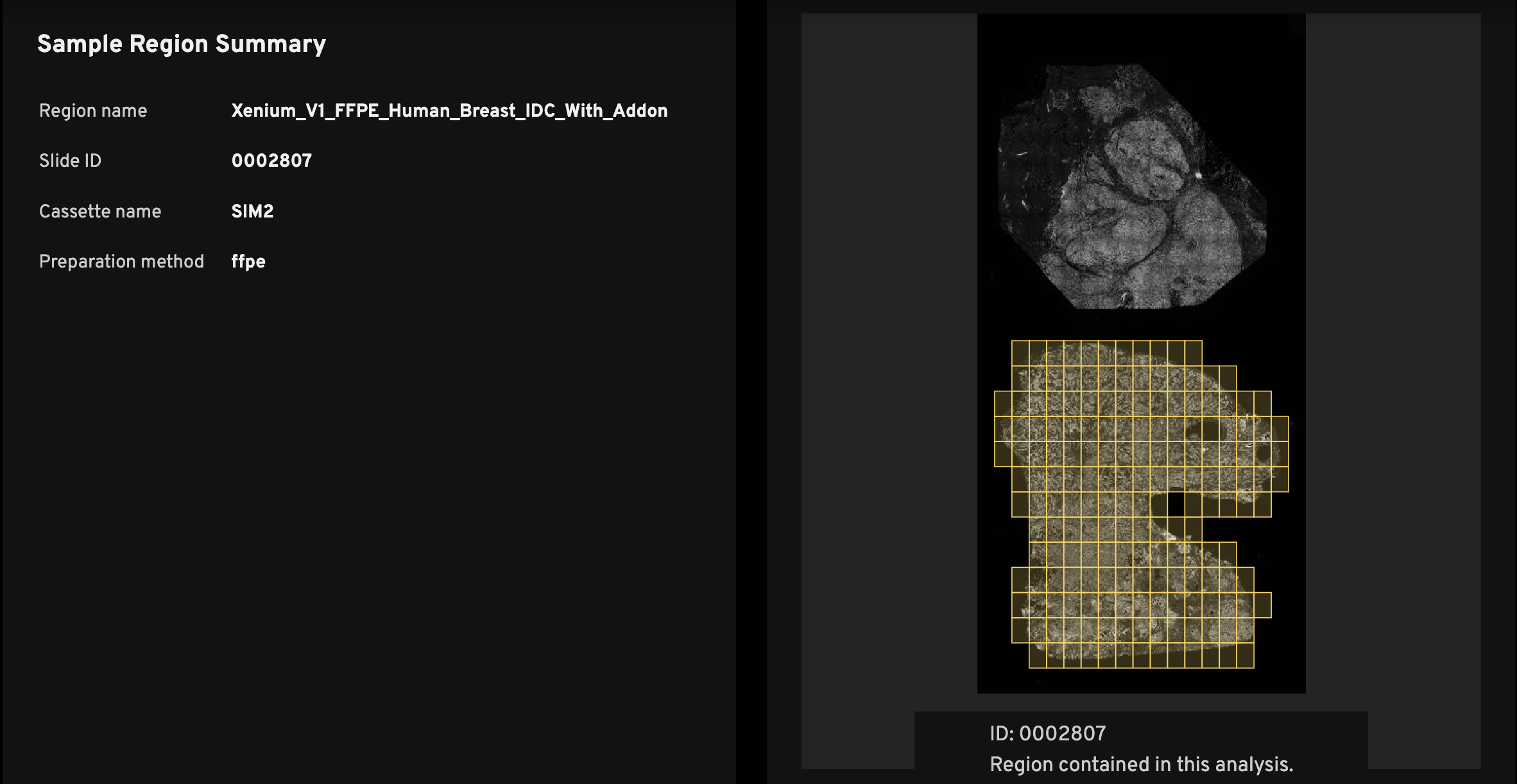
Every analysis_summary.html file has an overview scan (right), which shows the nuclei-stained (DAPI) image. The slide ID is displayed below the overview scan image (ID:). This ID location indicates the slide orientation, as it matches the relative location of the ID on the physical Xenium slide.
The purpose of the image is to help researchers quickly identify the tissue region on the slide that was used to generate the data in the Xenium output bundle. For example, if you have three mouse brain replicates on a slide, the overview scan quickly shows which among the top, middle, or bottom replicates the summary metrics correspond to.
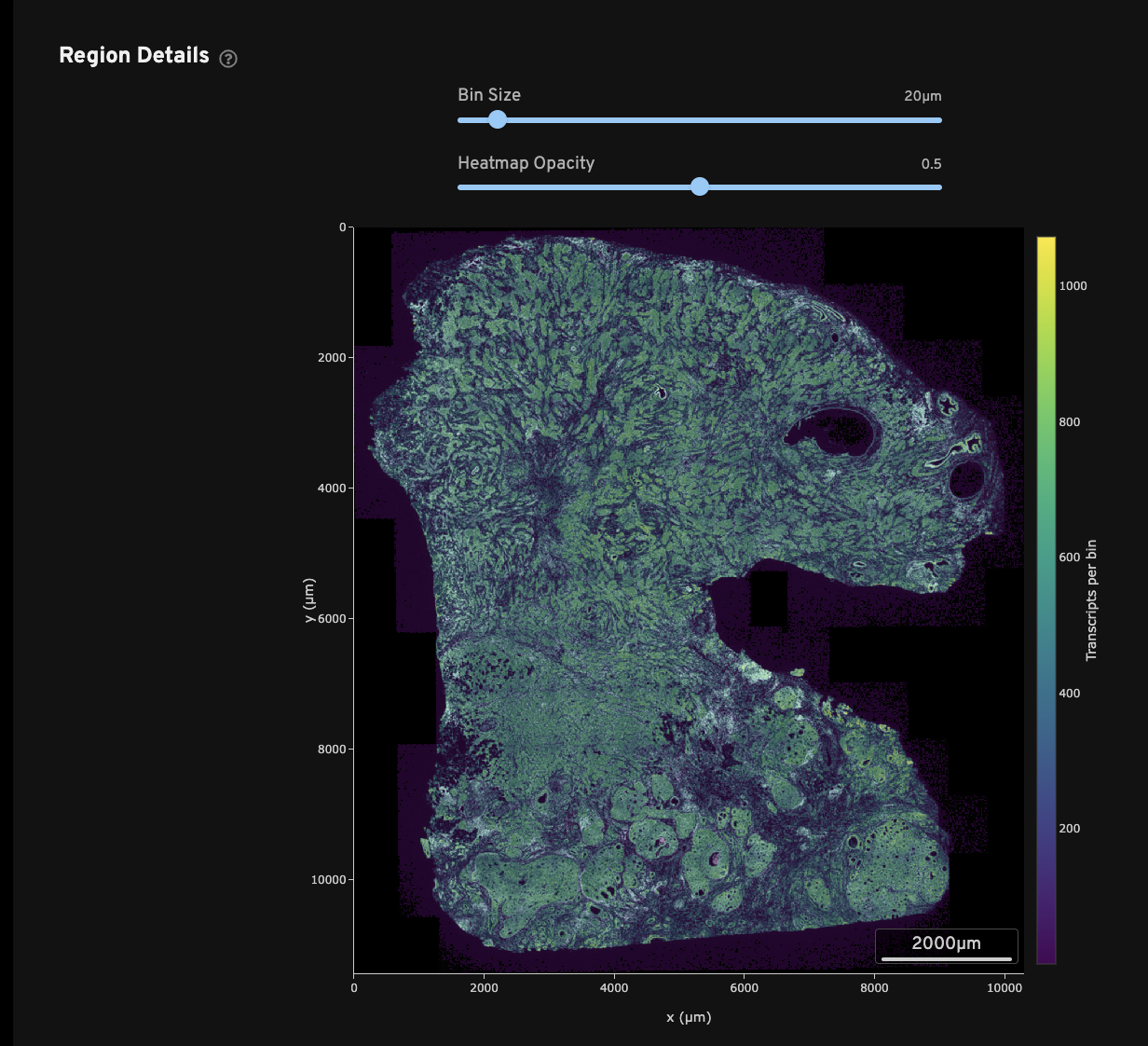
The Region Details view shows a DAPI image of the analyzed region along with a colored overlay of counts of high-quality decoded transcripts per bin. The overlay can be adjusted to various bin sizes and opacity scales as needed. Finer spatial resolution of decoded transcript density is shown with smaller bin sizes. If moused over, a zoomed-in DAPI image will be shown.
This view can be used to QC a variety of common sample preparation issues:
- Does the DAPI morphology image look as expected?
- Were there any tears in the tissue or detachment of tissues from the slide?
There are other questions that can be addressed using this view that require further experimental context, such as knowledge of the sample type. For example, does the general transcript density match up with the tissue morphology? If the tissue is a mouse brain, one would expect to see particularly high density in the hippocampus.
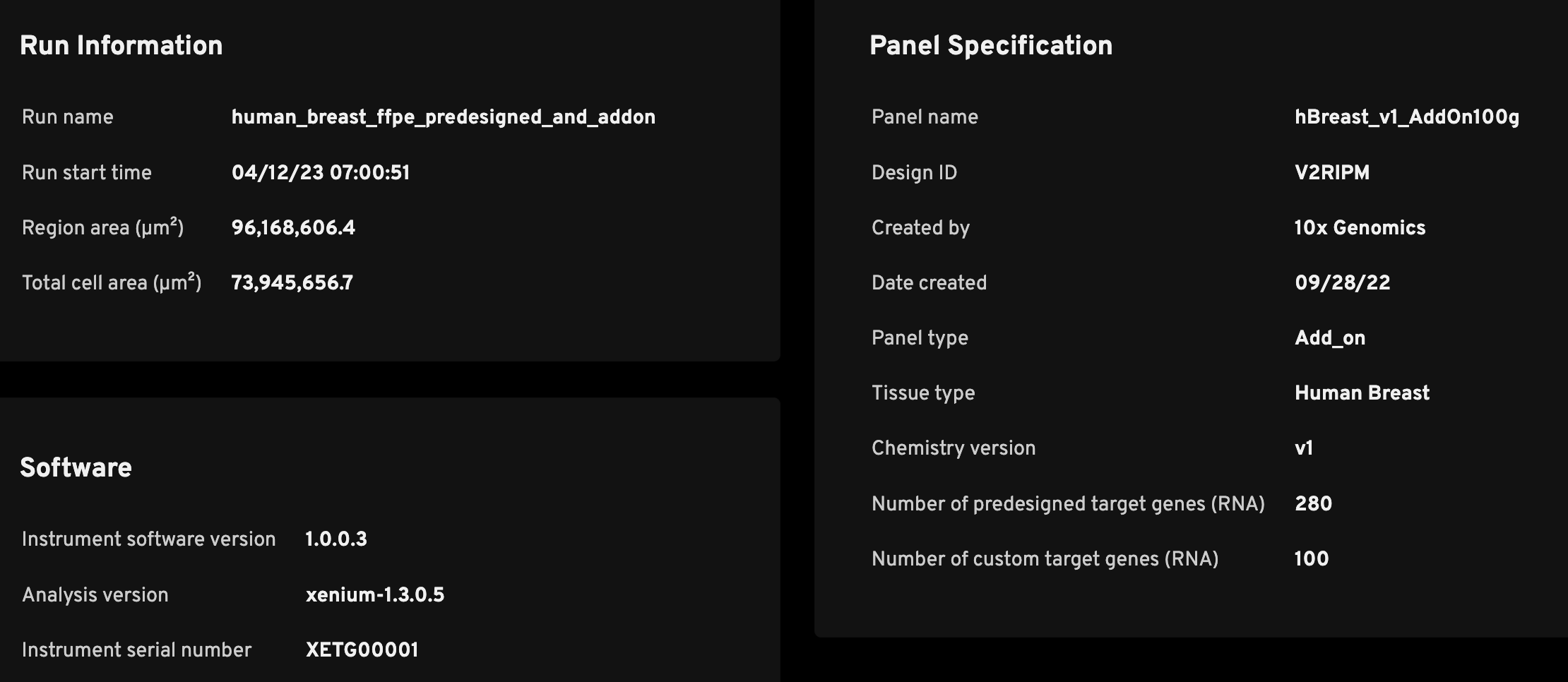
The Run Information section is meant to help researchers quickly determine key metadata regarding the run.
| Metric | Description |
|---|---|
| Run name | A unique identifier that was input in the instrument during the Initialize Instrument step (see the Xenium Analyzer User Guide). |
| Run start time | The time recorded by the Xenium Analyzer instrument (in UTC). |
| Region area (µm2) | The total area of imaged field of views (FOVs); the area of each FOV with tissue is summed. |
| Total cell area (µm2) | The summed area of detected cells; used for calculating transcript density (Decoded transcripts per 100 µm2). |
The Software section shows key metadata for troubleshooting purposes. 10x Genomics continually updates the instrument and analysis software on the Xenium instrument to introduce new features, fix bugs, and provide the best user experience.
| Metric | Description |
|---|---|
| Instrument software version | The version of firmware software used on the instrument (tracks instrument user interface changes as well). See release notes for updates. This instrument software version may not exactly match the analysis version. For example, the instrument software may be updated, but if no changes are added to the analysis software, the analysis version will not change. |
| Analysis version | The version of the Xenium Onboard Analysis pipeline that performs cell segmentation, decoding, and secondary analysis, and generates output files, including this analysis_summary.html. See release notes for updates. |
| Instrument serial number | A unique ID that may be helpful for troubleshooting with 10x Support. |
The Panel Specification section displays the Xenium panel that was used for the experiment. If the QC metrics look drastically different from what is expected, it may be prudent to double-check that the correct panel was used. The source of the metadata is the gene_panel.json file. If running a pre-designed panel, the JSON is already on the instrument. If running a custom or add-on panel, the JSON file would be produced by the Xenium Panel Designer or 10x support team and loaded onto the instrument during instrument initialization using a USB drive (see the Xenium Analyzer User Guide).
| Metric | Description |
|---|---|
| Panel name | The human-readable panel name. |
| Design ID | A unique string ID for the panel. |
| Created by | Indicates who designed the panel. |
| Panel type | Specifies whether the panel is pre-designed, add-on, or fully custom. |
| Tissue type | Indicates the species and tissue type. |
| Chemistry version | The assay chemistry version. |
| Number of target genes (RNA) | The number of genes targeted by the panel. For add-on panels, this metric is split between the number of pre-designed target genes and custom target genes. |
If the data were analyzed with Xenium Ranger, a new panel will display information about the analysis:
- Run ID
- Run timestamp
- Xenium Ranger software version
- The command(s) used to analyze the data
The Xenium codebook contains a collection of codewords that are assigned to genes in a gene panel. The pipeline uses the gene_panel.json to specify a given gene name to an indexed codeword. Each codeword is defined based on a pattern of fluorescent signal intensities recorded across channels and cycles. Some codewords are reserved for negative controls. For more information about transcript decoding, see the decoding, quality scores, and controls section of the Xenium algorithms page.
The Decoding Yield section provides QC metrics at a glance. Biological and experimental context may be needed for interpretation.
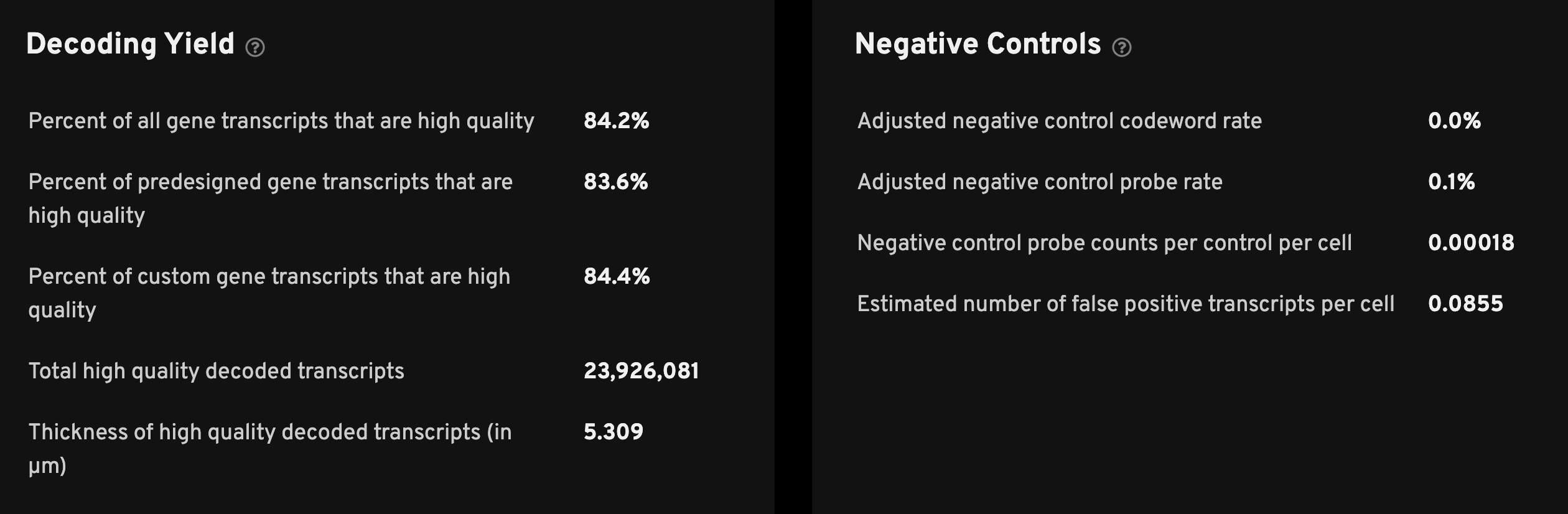
| Metric | Description |
|---|---|
| Percent of all gene transcripts that are high quality | The percent of transcripts from all genes that decode with high quality (≥ Q20). For experiments with custom panels, separate metrics are shown for pre-designed and custom genes since QC of custom genes is likely to be particularly important. |
| Total high quality decoded transcripts | The total number of decoded gene transcripts that were decoded with high quality (≥ Q20). |
| Thickness of high quality decoded transcripts (in µm) | The width in Z of high-quality transcripts measured in microns. The width is calculated as the difference of the 95th and 5th percentiles averaged over all the acquired fields of view (FOV). We exclude FOVs with fewer than 1000 high-quality transcripts before computing the average. This number typically ranges between 3-10 µm in tissues that are sliced at 5-10 µm. This number is proportional to the physical thickness of the tissue that is input into the assay for the same tissue type. The median high-quality transcripts per cell can be lowered when the measured thickness is lower than expected. When the tissue thickness is higher than expected the decoding quality scores may be affected. |
Below are descriptions for the Negative Controls metrics. The adjusted negative control probe rate measures both erroneous decoding and off-target binding, so it will always be greater than or equal to the adjusted negative control codeword rate. Both depend on the total number of high-quality transcripts detected, so a high value may result from lower detection of gene transcripts rather than nonspecific binding. For more information about negative controls, see the decoding, quality scores, and controls section of the Xenium algorithms page.
| Metric | Description |
|---|---|
| Adjusted negative control codeword rate | The estimated rate of false positives caused by erroneous decoding among high-quality (≥ Q20) transcripts. Estimated using negative control codewords such that any decoding to these codewords is definitively erroneous, and adjusted to estimate the rate of errors amongst all codewords. The rate is calculated as the fraction of high-quality transcripts that were assigned to negative control codewords, divided by the fraction of codewords in the panel that are negative control codewords. |
| Adjusted negative control probe rate | The estimated rate of false positive transcript signal caused by erroneous decoding and off-target binding among high-quality (≥ Q20) transcripts. Estimated using negative control probes that should not bind to any transcript sequence present in the tissue, and adjusted to estimate the rate of errors amongst all probes. The rate is calculated as the fraction of high-quality transcripts that were assigned to negative control probes, divided by the fraction of probe-associated codewords in the panel that belong to negative control probes. |
| Negative control probe counts per control per cell | This is the mean number of high-quality transcripts (Q-Score ≥ 20) assigned to cells that were decoded as negative control probes, which are probes that should not bind to any transcript sequence present in the tissue. The mean is taken across all cells and all negative control probes. More on metric calculations below. (Provided in Xenium Onboard Analysis v1.6 and later) |
| Estimated number of false positive transcripts per cell | This is the estimated mean number of high-quality transcripts per cell that do not represent true expression ("false positives"). It is estimated using the "Negative control probe counts per control per cell" metric and adjusted to estimate the number of false positives amongst all gene probes. It should be considered in the context of the total number of transcripts expressed per cell. (Provided in Xenium Onboard Analysis v1.6 and later) |
The "Negative control probe counts per control per cell" metric is derived first from calculating the absolute number of negative control probe counts and then calculating the metric per control probe per cell. The "Estimated number of false positive transcripts per cell" is derived from this metric. Both metrics are available in outputs generated with Xenium Onboard Analysis v1.6 and later. Derived values may differ slightly from those reported in summary outputs due to using rounded values. They are calculated using the following inputs:
Total high quality decoding transcripts: displayed on the Summary and Decoding tabs, includes genes (Q-Score ≥ 20) and negative controlsPercent of transcripts within cells: displayed on the Cell Segmentation tabAdjusted negative control probe rate: displayed on the Decoding tabNumber of target genes: the sum of genes in the pre-designed panel and any add-on custom genes, displayed on the Summary tabNumber of negative control probes in the panel: each pre-designed and add-on panel contains 20 negative control targets (except runs that use the Xenium Mouse Brain base panel, which have 27 negative control targets).Number of cells detected: hero metric on the Summary tab
- The Negative control probe count is calculated as:
where Q20 decoded transcripts in cells is calculated as:
- The
Negative control probe count per control per cellis then calculated as:
- The
Estimated number of false positive transcripts per cellis calculated as:
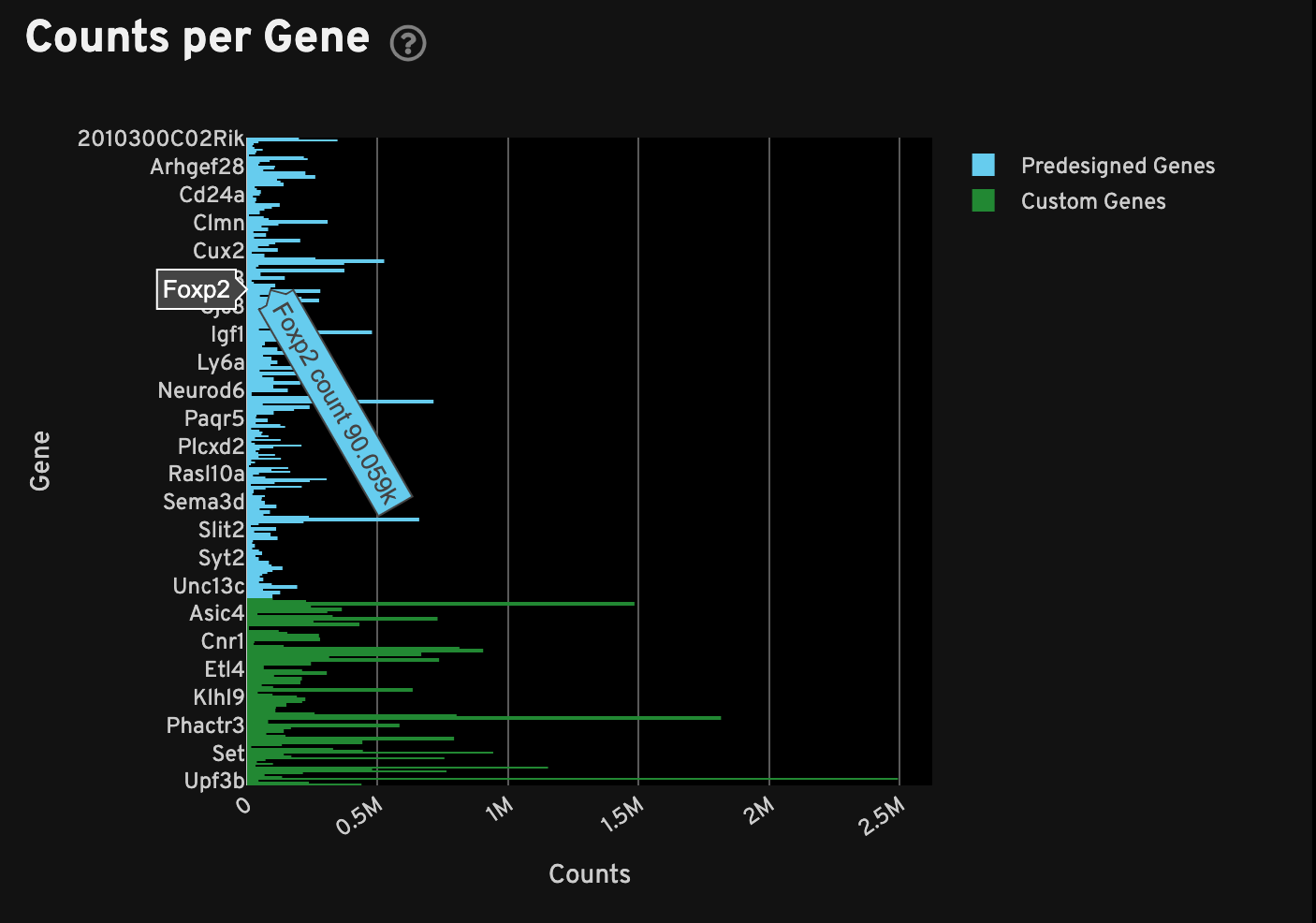
The Counts per Gene plot shows the total number of transcripts passing quality thresholds for every gene in the panel. This plot is a histogram of absolute counts of decoded transcripts (x-axis) by gene (y-axis). Genes are listed in alphabetical order. Hover over each bar to see which gene it corresponds to. This plot can be used to quickly compare multiple samples analyzed with the same panel, or to quickly compare add-on vs. pre-designed panels.
The Transcript Quality plot shows the distribution of quality scores for decoded transcripts, separating out transcripts that decode to panel genes, negative control probes, negative control codewords, and unassigned codewords. The transcript counts (y-axis) can be viewed in linear or log scale. High-quality samples should have panel genes with high quality scores (≥ 20) and negative controls of both types with low quality scores.
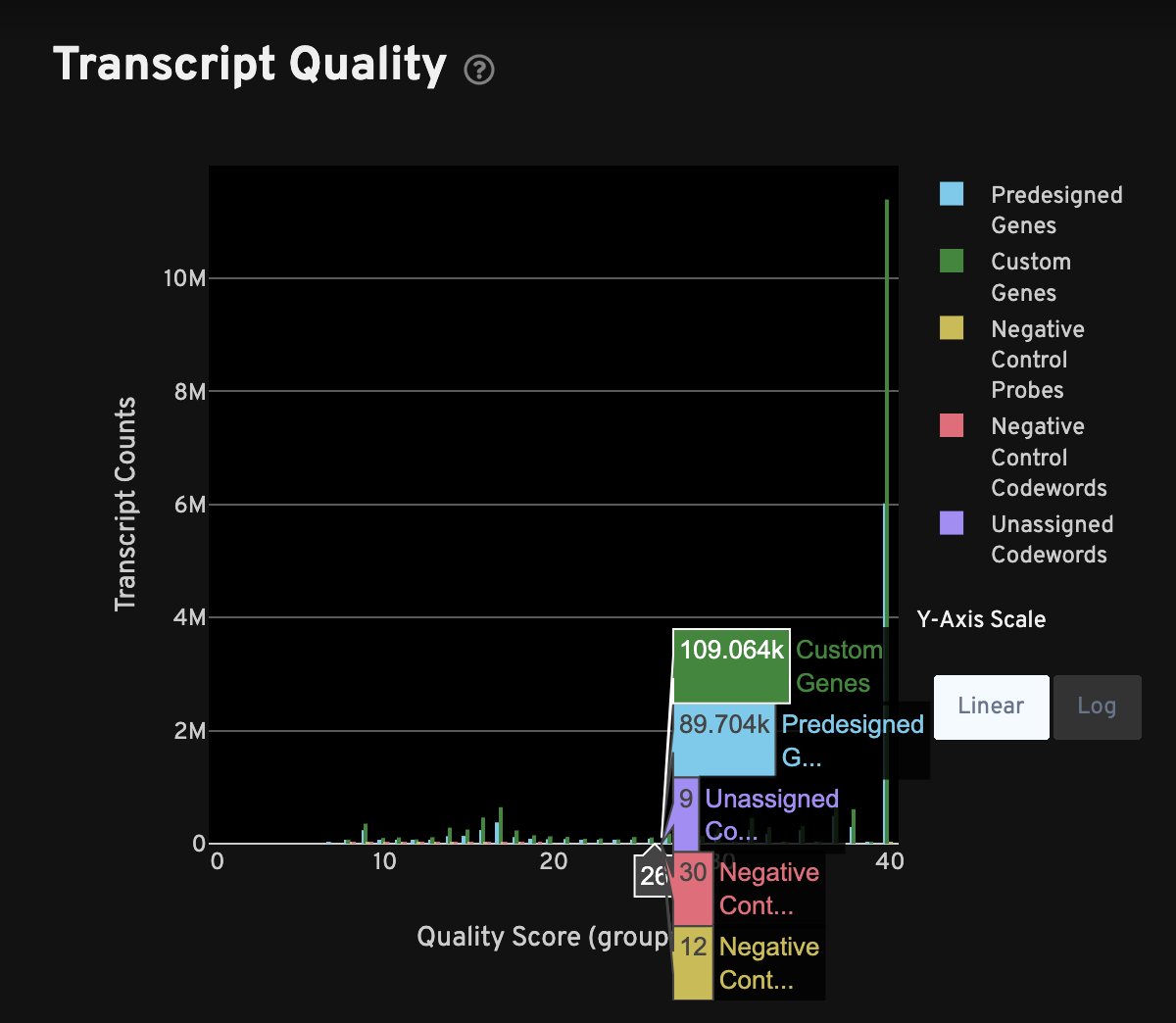
Click on a gene or negative control in the legend to hide it from view. This plot can be used to ascertain how much data is lost due to quality filtering. Data with quality scores < 20 are filtered from the cell-feature matrix, and downstream analyses using the matrix file, but these filtered transcripts can still be found in the transcripts files. For more information, see Understanding Xenium Outputs.
For each gene, the Gene-Specific Transcript Quality plot shows the total number of decoded transcripts of any quality (x-axis) against the mean quality of those decoded transcripts (y-axis). The x-axis (total transcripts per gene) is plotted on a log10 scale, while the y-axis is on a linear scale.
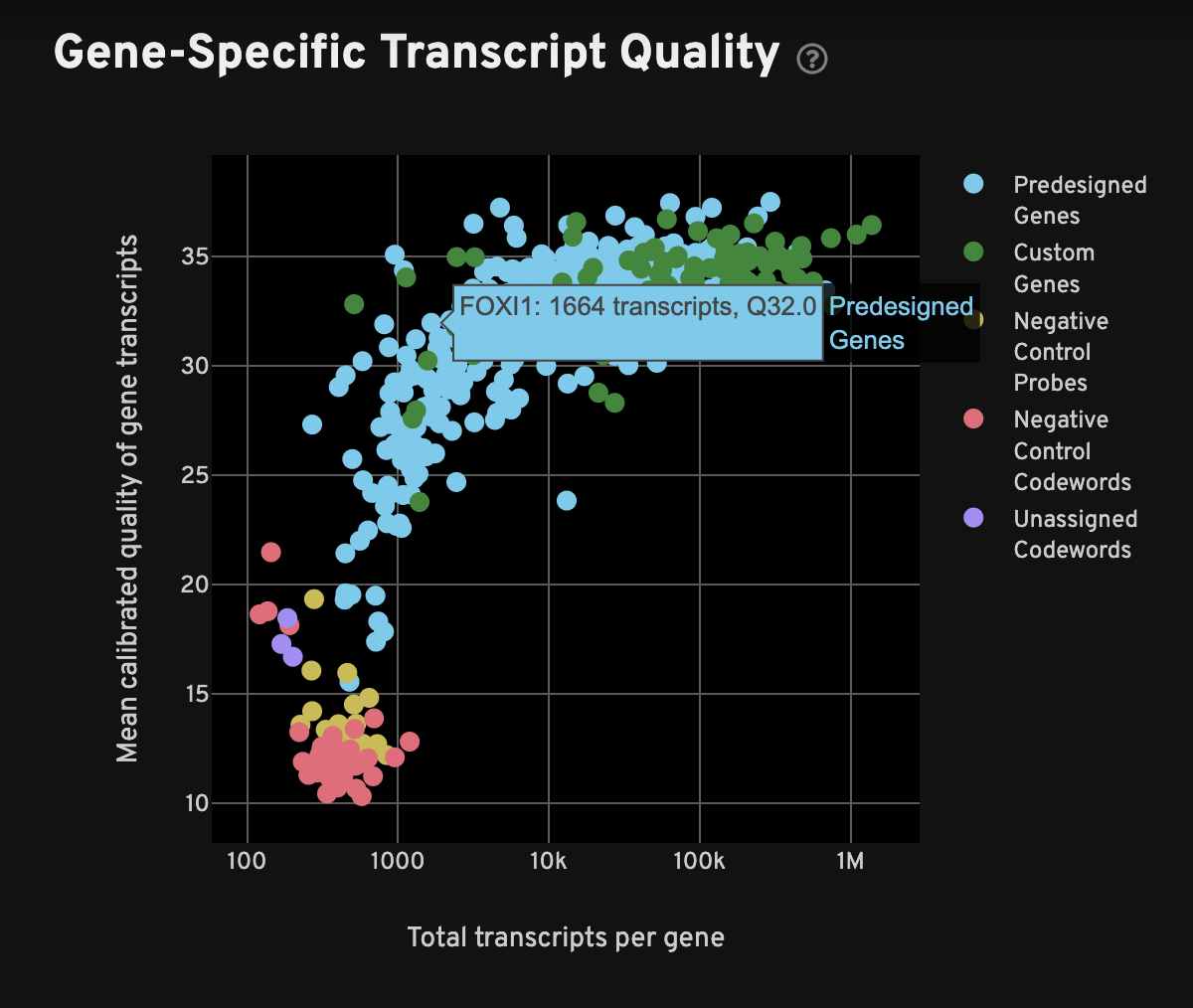
When interpreting this plot, one would typically expect to see all or most genes have mean quality scores ≥ 20, and the controls should be < 20; in other words, genes should be at the top-right quadrant and controls should be in the bottom-left quadrant. Examine genes, especially custom genes, in the panel for any performance problems (i.e., low density or low mean Q-Score may indicate problems in the performance of a given gene).
Negative controls for both decoding and probes should have low transcript counts and low quality:
- A high rate of negative control codewords may indicate possible imaging issues, such as autofluorescence in one or more cycle-channels that are part of a negative control codeword. Check the RNA images in the
aux_outputs/directory. - High quality and count of all negative control probes may indicate assay workflow issues, such as nonspecific probe hybridization or ligation conditions. It is ok if only a few negative control probes show high quality and/or count.
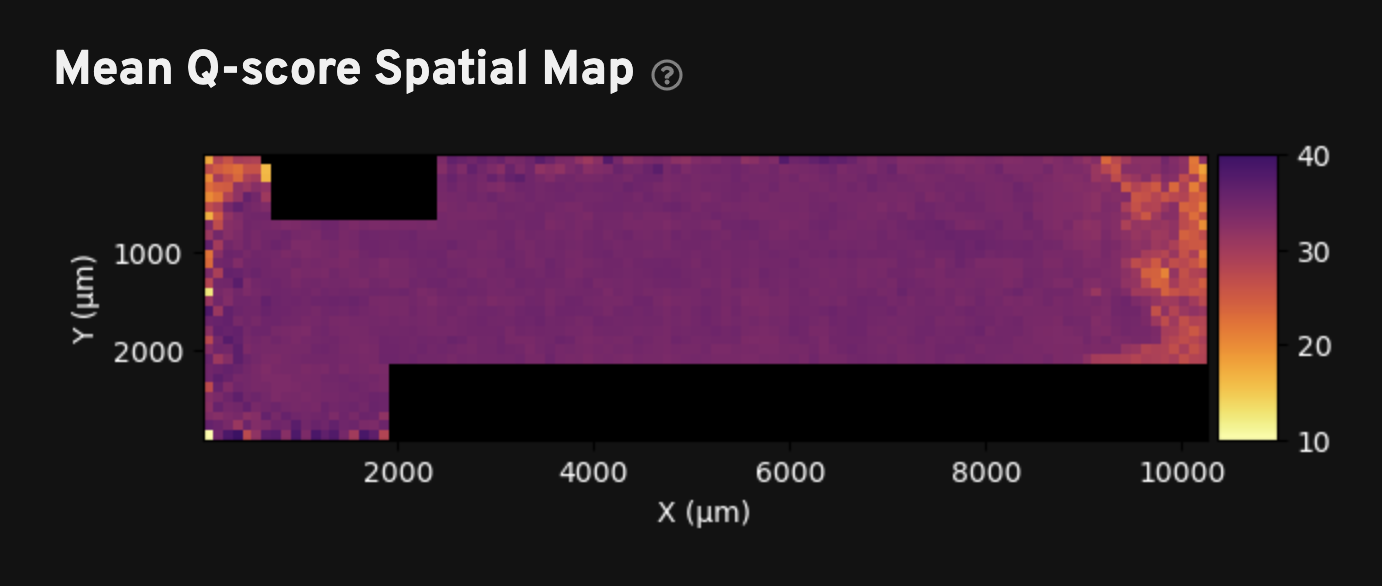
The Mean Q-score Spatial Map plot shows binned transcript quality scores across the sample area. This plot can be used to identify any technical artifacts associated with one or more fields of view (FOVs). The example screenshot below is from a pancreatic cancer dataset:
The purpose of cell segmentation is to approximate cell boundaries so that transcripts can be assigned to cells. Downstream, these results will be used to produce a cell-feature matrix, similar to those output by existing single cell and spatial technologies. For more information, see the cell segmentation section of the Xenium algorithms page.

Here are descriptions for the Segmentation Metrics:
| Metric | Description |
|---|---|
| Fraction of empty cells | The percentage of all cells without decoded high-quality transcripts. This should typically be a low value but an acceptable range for each region is dependent on the gene panel and tissue sample. An error message will appear at the top of the analysis summary if the percentage is greater than 10%. |
| Percent of transcripts within cells | Percent of high-quality transcripts that are found within cells. A warning message will appear at the top of the analysis summary if the percentage is less than 50%. Low values can be caused by underdetection of cells or sample preparation issues leading to mis-localized transcripts. Unassigned transcripts are excluded from the cell-feature matrix. |
| Cells per 100 µm2 | The density of cells per 100 microns squared. This metric may vary by tissue type and cell size. |
| Median genes per cell | The median number of unique genes detected per cell. Cells with zero transcripts are excluded from the calculation. For add-on panels, this metric is also split between median pre-designed genes or custom genes per cell. |
| Median transcripts per cell | The median number of transcripts per cell. Cells with zero transcripts are excluded from the calculation. For add-on panels, this metric is also split between median pre-designed or custom transcripts per cell. |
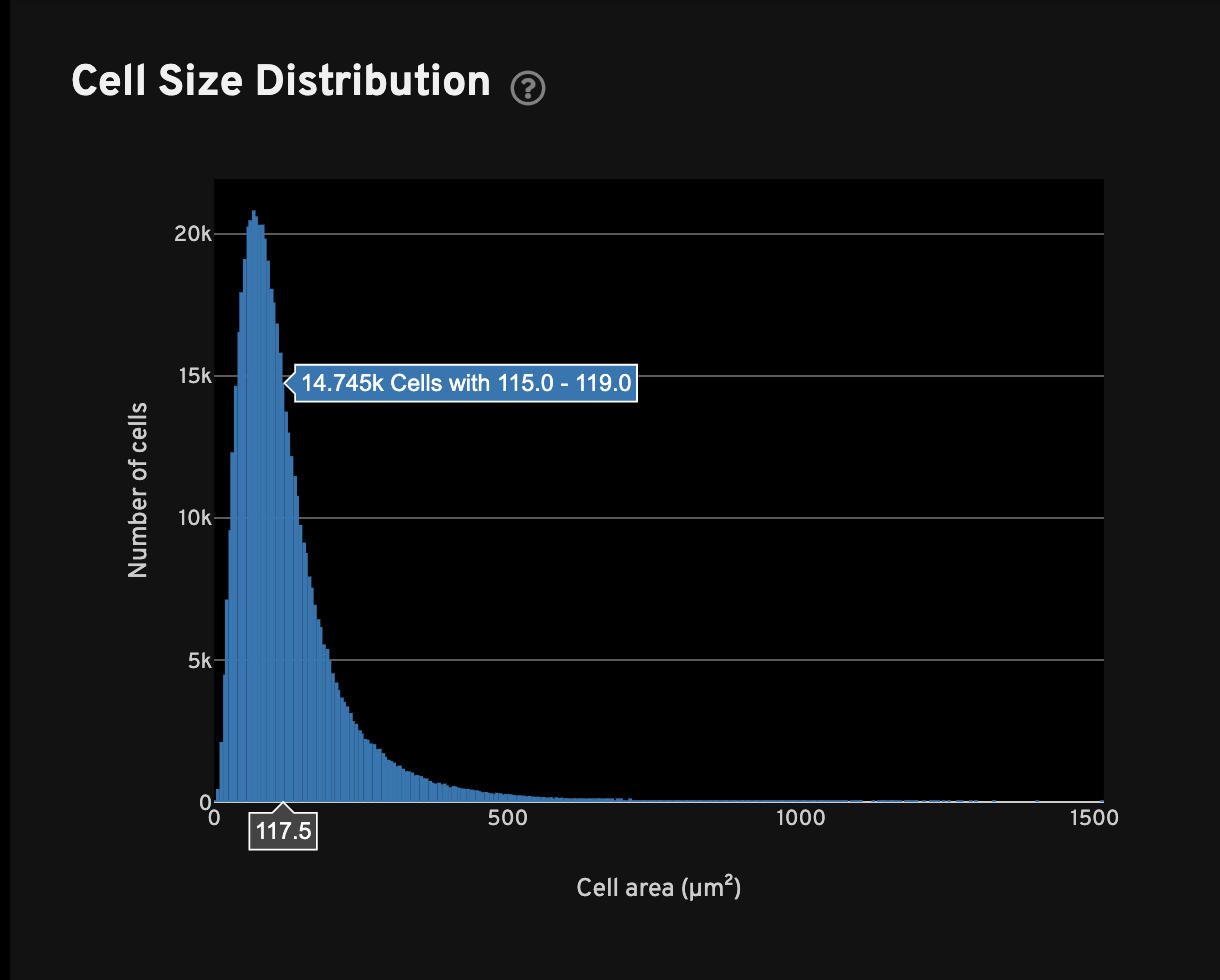
For all cells with transcripts, the Cell Size Distribution view shows a histogram of cell area in µm2. The area is computed from the cell segmentation mask.
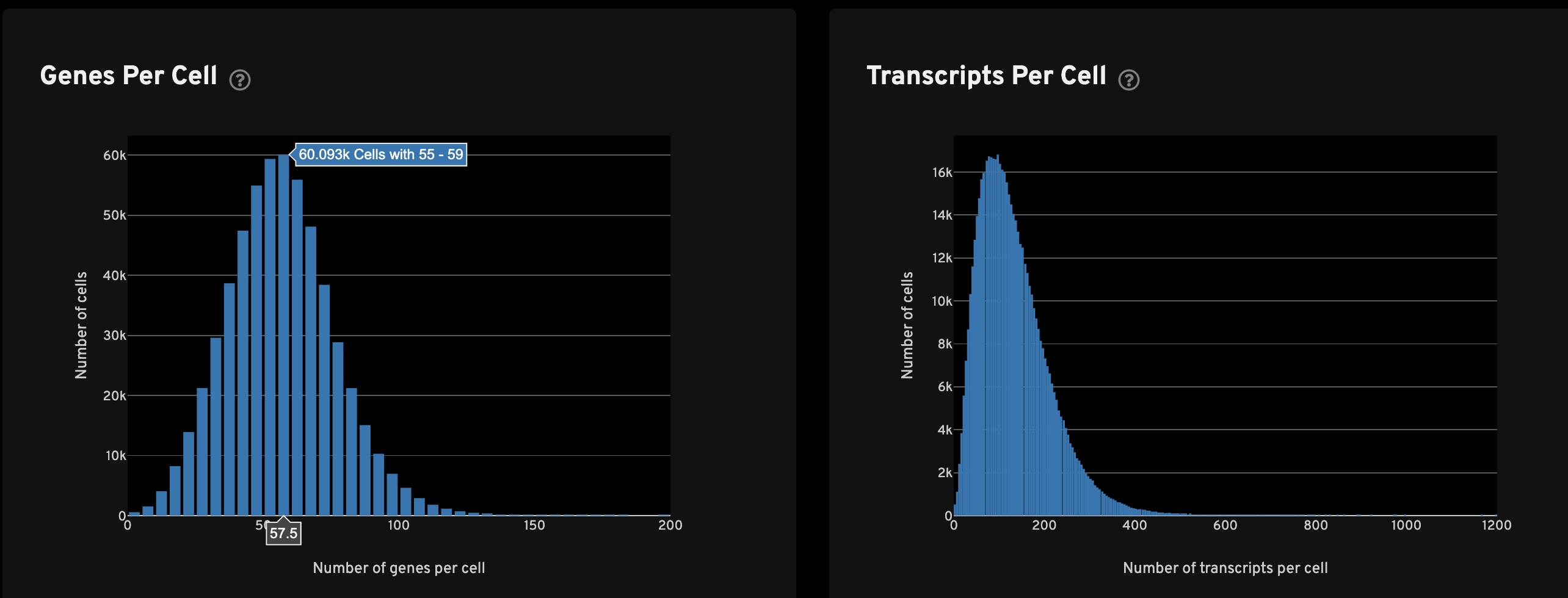
The Genes per cell view shows a histogram of the total number of unique, panel (non-control) genes found in each cell for all cells with transcripts. The Transcripts per cell view shows a histogram of the total number of transcripts found in each cell over all panel (non-control) genes for all cells with transcripts.
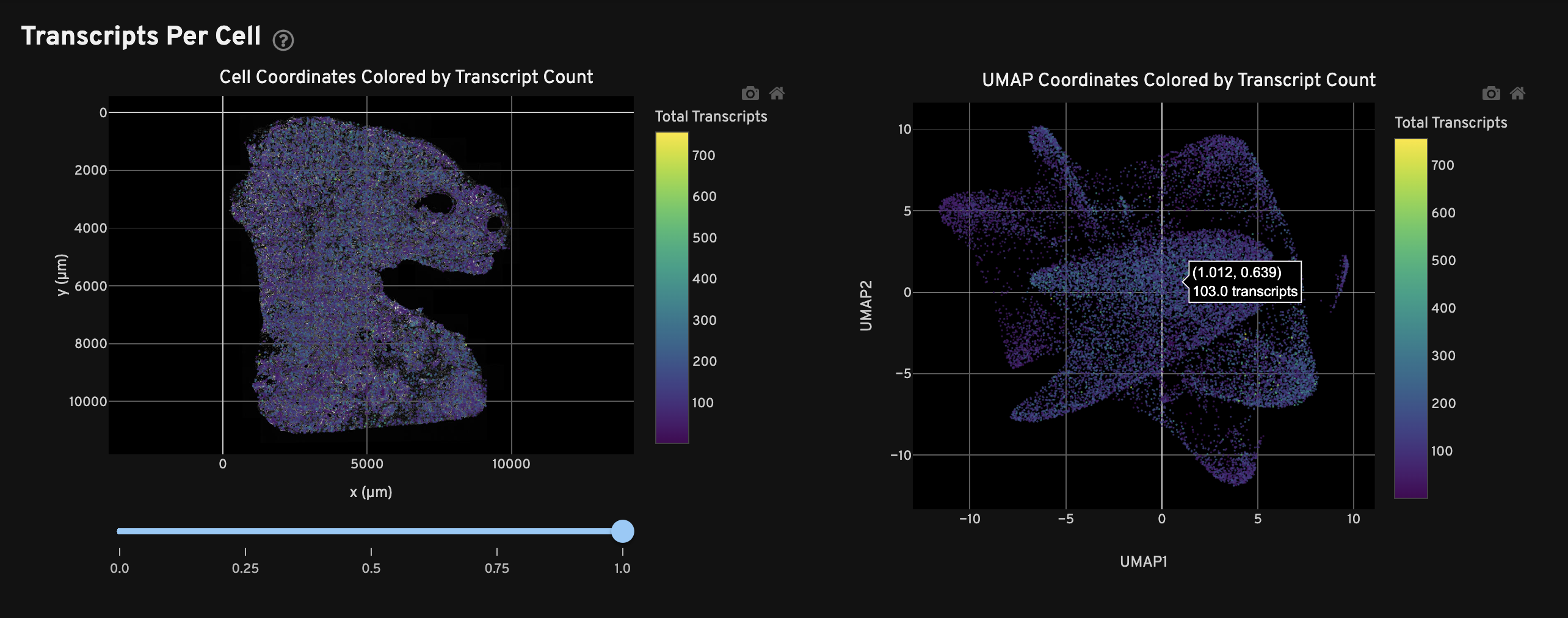
The Transcripts Per Cell view shows the spatial distribution (left) and UMAP projection (right) of cells colored by the total number of transcripts detected in each cell. For very large samples, a subset of cells may be plotted for better visualization performance.
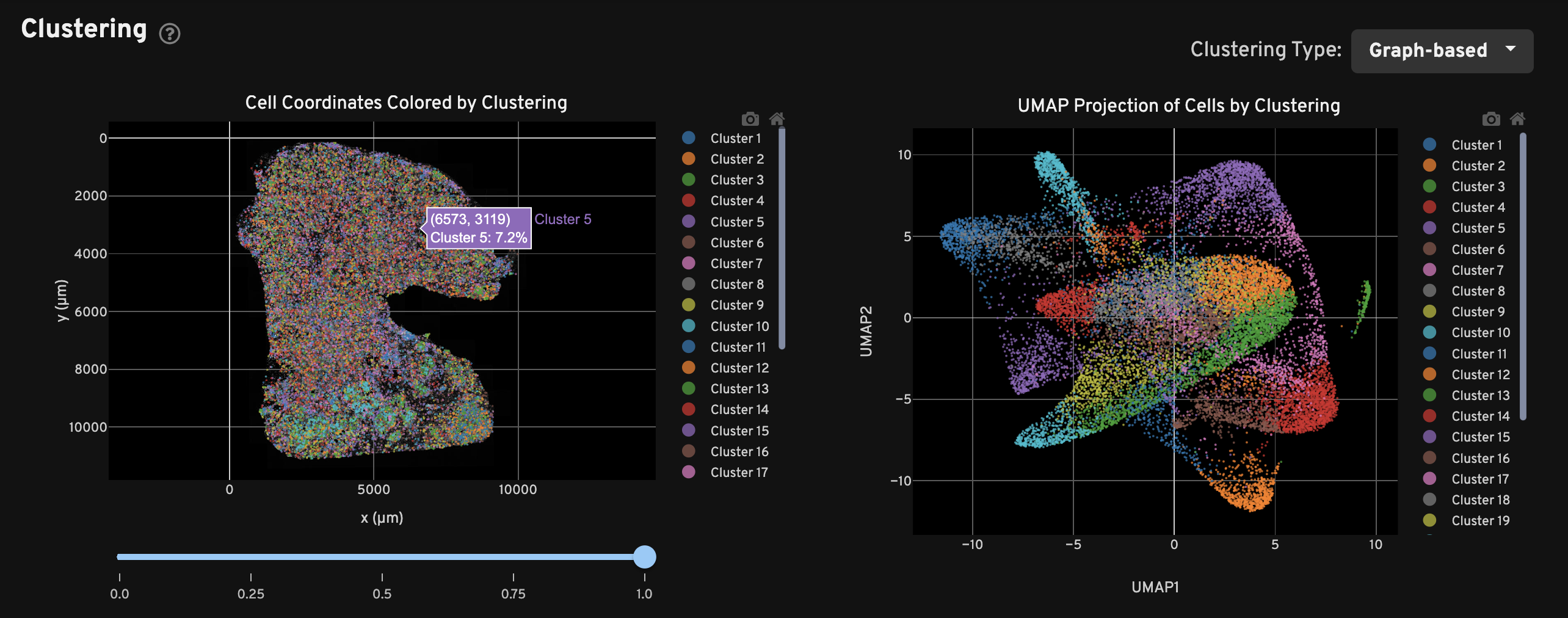
The Clustering view shows the spatial distribution (left) and UMAP projection (right) of cells colored by cluster assignment using the Onboard Analysis pipeline's automated clustering algorithm. The clusters should reflect groups of cells that have similar expression profiles. In the left plot, cells are colored according to their cluster assignment and plotted in their spatial location. Only cells with a nucleus detected by the DAPI stain are used in the clustering algorithm.
In the right plot, the axes correspond to the 2-dimensional embedding produced by the UMAP algorithm. In this space, pairs of cells that are close to each other have more similar gene expression profiles than cells that are distant from each other.
The Top Features by Cluster table displays results from the Onboard Analysis pipeline's automated differential expression analysis. For each cluster, the table shows features that are more highly expressed in that cluster relative to the rest of the sample.
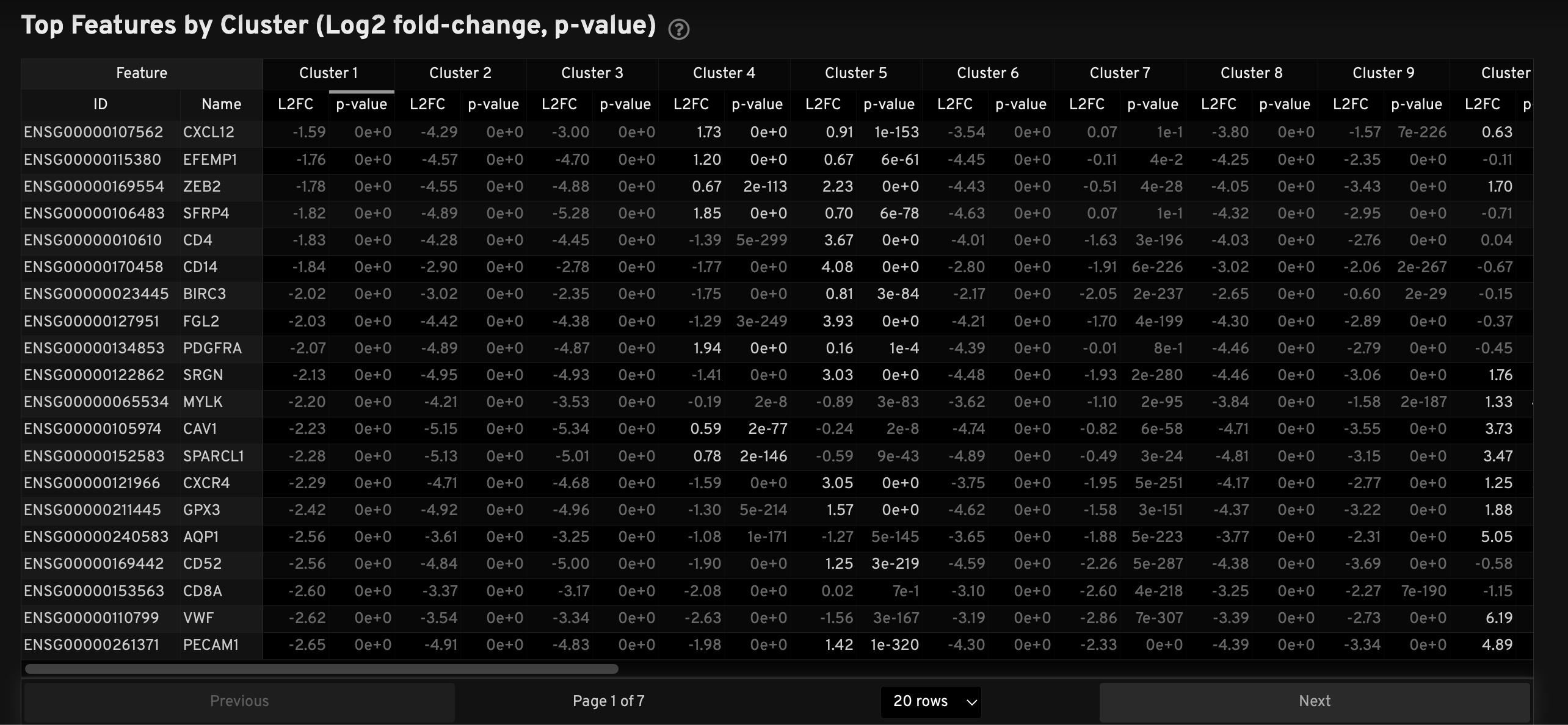
A differential expression test was performed between each cluster and the rest of the sample for each feature.
- The Log2 fold-change (L2FC) is an estimate of the log2 ratio of expression in a cluster to that in all other cells. A value of 1.0 indicates a 2-fold greater expression in the cluster of interest.
- The p-value is a measure of the statistical significance of the expression difference and is based on a negative binomial test. The p-value reported here has been adjusted for multiple testing via the Benjamini-Hochberg procedure.
In this table, you can click on a column to sort by L2FC or p-value for each cluster. Features were filtered (mean object counts > 1.0) and the top N features by L2FC were retained for each cluster. Features with L2FC < 0 or adjusted p-value ≥ 0.10 are grayed out. The number of top features shown per cluster, N, is set to limit the number of table entries shown to 10,000 (N=10,000/K2 where K is the number of clusters). N can range from 1 to 50. For the full table, please refer to the differential expression CSV files produced by the pipeline.
The gallery in this tab shows a stitched and reconstructed fluorescence image of the analyzed region obtained by downsampling and processing the internal sensor data for each cycle (row) and channel (column).
The intensity values in the thumbnail image give the average fluorescence intensity measured by the sensor in units of photoelectrons (pe) per square micron of tissue area. They are downsampled to 200 x 200 pixels from the high-resolution images available in aux_outputs/per_cycle_channel_images/.
This gallery can be useful for quick data QC, such as checking for stitching errors, total cycle dropouts or oversaturation, debris, or low transcript density areas. Patterned changes in fluorescence intensities across cycle-channel plots are a signal of good image quality. For example, in the images below, certain tissue areas are dim in some cycles and bright in others:
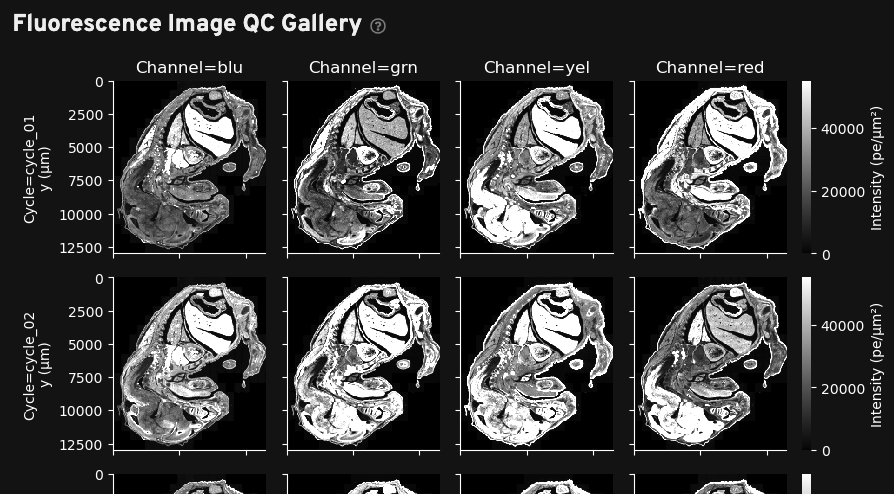
Fluorescence signals that are always high in the same location in all cycles indicate tissue autofluorescence and may affect transcript data at that location.