Single cell RNA-seq: An introductory overview and tools for getting started
Single cell RNA sequencing (scRNA-seq) measures whole transcriptome gene expression in individual cells, offering a detailed view of how cells function and interact within complex tissues. This blog introduces the benefits of this experimental approach as well as a basic overview of how our Chromium scRNA-seq assays work, and how you can get started.

A beginner’s guide to single cell RNA sequencing
What is single cell RNA-seq? What can it do for my research? Dive into this interactive web portal to find out more about single cell technology, explore powerful applications across research areas, and get answers to common questions about cost, sample compatibility, workflow complexity, and data analysis.
Swimming in a sea of cells: Why single cell RNA-seq?
Complex biological systems result from the coordinated functions of individual cells performing an intricate "dance," each contributing their own special role to the ensemble. Because of this complexity, research and analysis of genes in organisms, tissues, or cell populations is often limited by traditional bulk RNA-seq methods. As scientists, we intuitively understand that when we "grind and find" in order to perform experiments on our diluted mix of RNA, we are really only getting a small idea of what might be going on in our cells of interest: cell-to-cell differences are missed and cellular heterogeneity can be completely masked. In fact, bulk RNA expression analysis often describes an inferred state in which none (or very few) of the cells actually exist!
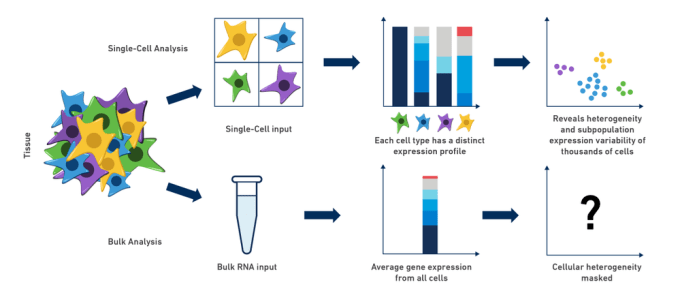
Perhaps you are a neuroscientist, working in the mouse brain, studying the development of a subpopulation of neurons that produce a specific neurotransmitter. If only you could isolate that population and analyze it on a cell-by-cell basis, the potential insights would be, indeed, neuronally stimulating! Similarly, in the case of cancer research: how do you best tease apart gene expression and, perhaps more importantly, cellular heterogeneity in a tumor? What if you could compare not only inter-tumor, but intra-tumor differences and explore the immune cell populations related to the tumor microenvironment? This diversity, often obscured by bulk RNA-seq methods, can be revealed by using single cell RNA-seq to specifically explore these clinically significant cell populations. The examples of drawbacks to bulk analysis are abundant, and dissecting cell-type differences in most, if not all, of these complex biological systems is critical to our understanding of their contributions, especially during development and in the progression of disease.
How do we isolate and analyze single cells?
10x Genomics’ original single cell RNA-seq (scRNA-seq) technology, the Universal 3’ Gene Expression assay (previously known as the Chromium Single Cell 3’ solution) allows you to analyze transcriptomes on a cell-by-cell basis through the use of microfluidic partitioning to capture single cells and prepare barcoded, next-generation sequencing (NGS) cDNA libraries. Our Universal 5’ Gene Expression assay (previously known as Chromium Single Cell Immune Profiling) works in a similar way.
Specifically, single cells, reverse transcription (RT) reagents, Gel Beads containing barcoded oligonucleotides, and oil are combined on a microfluidic chip to form reaction vesicles called Gel Beads-in-emulsion, or GEMs. Each functional GEM contains a single cell, a single Gel Bead, and RT reagents. Within each GEM reaction vesicle, the single cell is lysed, the Gel Bead is dissolved to free the identically barcoded RT oligonucleotides into solution, and reverse transcription of polyadenylated mRNA occurs. As a result, all cDNAs from a single cell will have the same barcode, allowing the sequencing reads to be mapped back to their original single cells of origin. The preparation of NGS libraries from these barcoded cDNAs is then carried out in a highly efficient bulk reaction.
This process is now powered by GEM-X technology, which improved upon our previous Next GEM technology with enhanced reagents and microfluidics. The redesigned GEM-X microfluidic chips utilize the flow of partitioning oil to facilitate GEM formation. Twice as many GEMs are generated at smaller volumes, reducing multiplet rates two-fold and increasing throughput capabilities. Leveraging our Universal assay family, users can process 80K to 960K cells per kit in a single six-minute Chromium X instrument run with up to 80% cell recovery efficiency.
The video below provides a great visual explanation of how this all works.
Our newest Chromium scRNA-seq assay, Flex Gene Expression, also leverages the GEM-X technology foundation while allowing researchers to profile fresh, frozen, and fixed samples, including FFPE tissues and fixed whole blood, on a more flexible schedule. The Flex Gene Expression sample prep workflow involves generating a fixed single cell or nuclei suspension. Samples are permeabilized and hybridized with probe sets, then can proceed to partitioning into GEMs on the Chromium X instrument as described above.
The Flex Gene Expression assay provides highly sensitive protein-coding gene coverage for human or mouse samples, with options for customizable coverage to address specific experimental needs. Notably, Flex yields high-quality results from samples with low-quality or damaged RNA. This method also enables more flexible workflow options and sample-type compatibility. Quick and easy fixation protocols preserve biology while reducing urgency from sample processing and streamlining experimental logistics, ideal for longitudinal or multi-site studies, or when working with precious clinical samples. Moreover, Flex enables high scalability and cost efficiency, with a throughput capacity of 80,000 to 5.12 million cells per kit.
The video below* breaks down how the Flex assay works.
*This video features a Flex workflow that previously leveraged Next GEM technology. The Flex assay is now powered by GEM-X technology.
From sample prep to how-to videos: Helpful links for getting started
The Universal and Flex workflows begin with your cells of interest, followed by NGS library construction using our reagent kits and a Chromium X Series instrument. Preparing your barcoded cDNA library (as generally described in the videos above) is an important step, and true to the saying "garbage in, garbage out," preparing high-quality cell suspensions is key to getting the most out of your single cell RNA-seq data. If you need assistance with the cell preparation process, we have multiple demonstrated protocols available on our website, such as dissociation of neural tissue or preparation of moss protoplast suspensions for use in scRNA-seq, with more protocols being added regularly to our support website. For guidance and recommendations on how to best prep your samples once they are in suspension, a good starting point is our single cell preparation guide, where you can learn more about best practices and view a general protocol. If you are more of a visual learner, our how-to videos should help walk you through the sample and library preparation processes.



- Single Cell Prep Guide
- Demonstrated Protocols
- Chromium X Series Instruments
- User Guides (Universal 3’ Gene Expression)
- User Guides (Universal 5’ Gene Expression)
- User Guides (Flex Gene Expression)
- How-To Videos
- Cell Ranger Pipeline
- Loupe Browser Software
- Cloud Analysis
Analyzing your single cell data
After your library is prepared, sequencing can be performed with a number of different sequencing platforms, including Illumina, PacBio, Ultima Genomics, and Oxford Nanopore sequencing instruments. This leads to the fun part: gleaning biological insights from your single cell RNA-seq data! Advanced tools and software, such as the Cell Ranger pipeline, are essential to effectively process sequencing data and extract meaningful biological insights. The (potentially) massive amount of data you generate from Chromium scRNA-seq assays can be easily handled by our software and visualization tools, which were designed to take away a lot of the guesswork: Cell Ranger pipelines transform the barcoded sequencing data into files ready for single cell expression analysis, and visualization is accomplished via Loupe Browser software. If you would like a step-by-step overview, these tutorials on the software support site cover everything from transcript counting with Cell Ranger pipelines to beautiful data visualization with Loupe Browser software.
There have also been more than a thousand tools developed by the community for single cell RNA-seq analysis. We put together some tips for continuing your journey after running the Cell Ranger pipeline. Read more about data analysis topics on our Analysis Guides page, where we summarize important analysis topics, provide guides and tutorials, and share up-to-date info on all things 10x Genomics data analysis.
If you have any other questions or concerns about getting started with Chromium assays, feel free to contact us directly. Whether it be tracking brain organoid development, studying bone marrow transplantation in patients with acute myeloid leukemia, or unraveling a novel pathway for intestinal stem cell self-renewal, the number of Chromium Single Cell publications is growing daily, and we look forward to hearing about all of your amazing research!

Blog: Choosing the right single cell assay for your experiments
Ready to take the next step to explore options for single cell research? Read this blog for a breakdown of the single cell RNA-seq assay families on our Chromium platform, Flex and Universal. Explore their advantages, plus practical considerations for experimental setup, and find guidance for choosing the right single cell assay.
Editor’s Note: This post was originally published in 2017 and has been updated as of October 2025 in accordance with the current 10x Genomics style guidelines, accurate resource links, and information about new additions to the Chromium Single Cell platform. The original author was Sheila Clark, PhD.
