There are many scripts and packages in the single cell analysis ecosystem. You may want to import and export projections, categorical labels, gene lists and filters into and out of Loupe Browser. Options to import and export data in Loupe Browser are usually inside a menu that is displayed after clicking a button with three vertical dots.
The types of data and formats that can be imported and exported from Loupe Browser are described here.
In Loupe Browser 3.1.0 and later, you can import custom projections. This allows you to import a projection of the data using your favorite software packages like Seurat or Scanpy to compute UMAP, t-SNE, PCA, or MDS projections. You can also import layouts from trajectory inference methods.
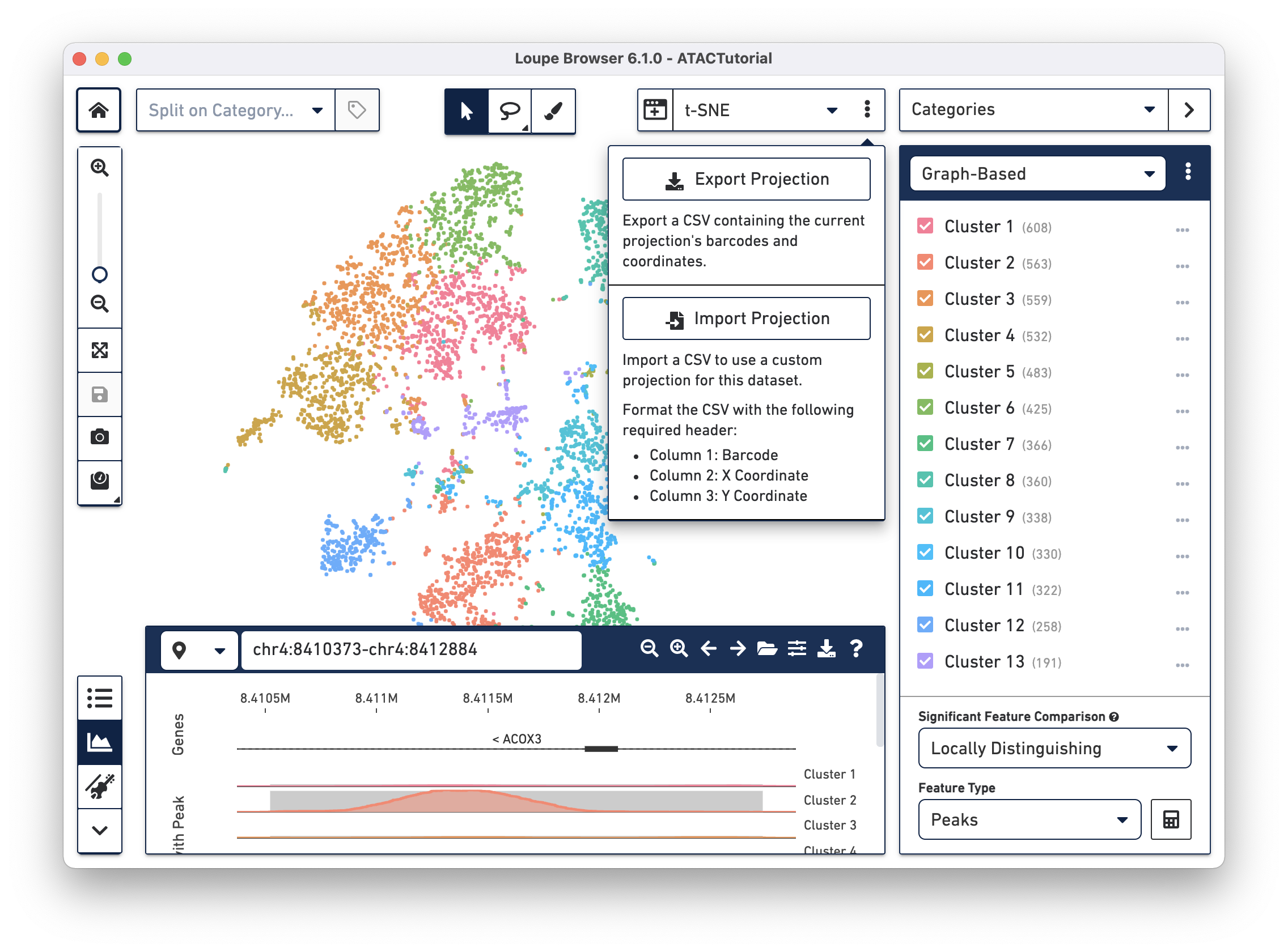
The CSV format for importing projections is:
- Header is required
- First column: barcodes that are at least a subset of the barcodes in the
.cloupefile - Second column: numbers which represents the X coordinates
- Third column: numbers which represents the Y coordinates
Here is a simple example for a small subset of barcodes from the bundled AMLTutorial dataset.
Barcode,UMAP-1,UMAP-2
AAACATACGGTACT-1,-10.18537,-5.062836
AAACATTGCTCGCT-1,-7.0603027,2.789417
AAACATTGCTTCTA-1,-8.413968,-3.3959498
AAACATTGGCGATT-1,-3.0396965,-7.3784895
AAACCGTGCCTACC-1,-7.6169457,2.5746179
To import new projections, click on the action button to the right of the projection selector. Once a custom projection is uploaded, you are also able to rename it or delete it.
Loupe Browser allows you to import and export categories. This allows you to integrate clustering data from other pipelines or scripts, or export manually assigned categories into custom scripts. To do so, when Categories mode is active, simply click on the action button to the right of the active category name, and select from one of the import/export options.
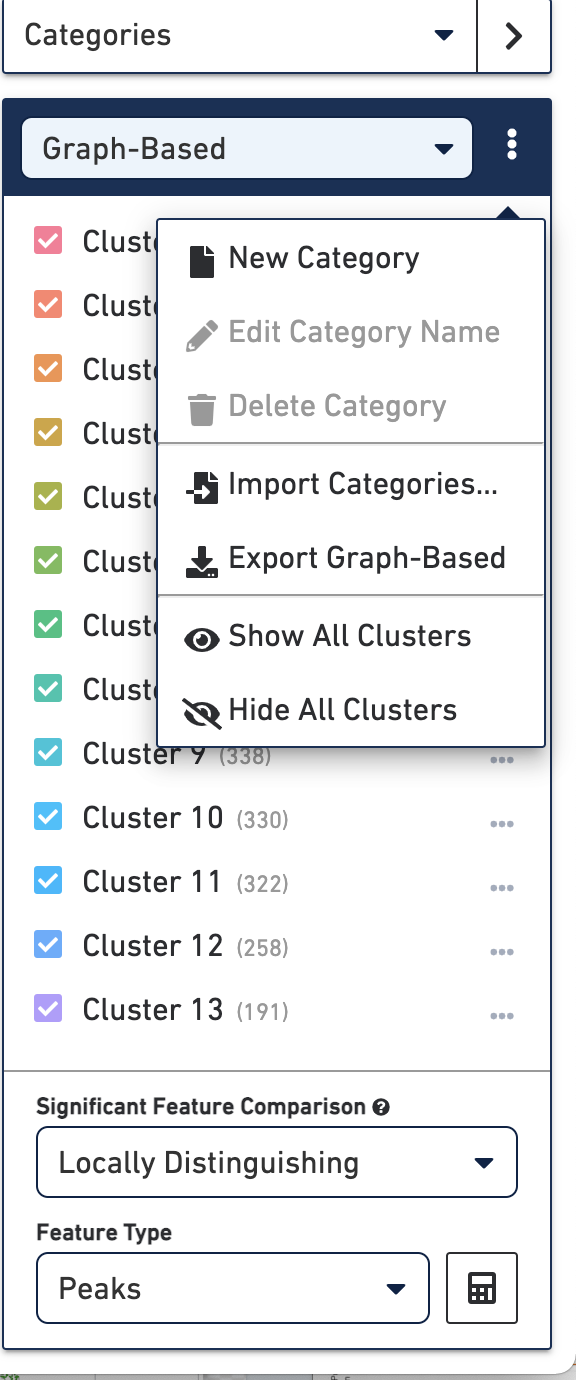
The format to create a categories CSV file is:
- First row must be a header
- A column header must be
barcodeand the column's contents must match at least a subset of the barcodes in the.cloupefile - Addition column headers are category names and the column's contents are cluster labels.
barcode,id
AAACATACAGTACC-1,Monocytes
AAACCGTGTCGTGA-1,Monocytes
AAACATACCATTCT-1,Mature Ery
AAACATACTCGCAA-1,Mature Ery
AAACCGTGGCCATA-1,Mature Ery
In this case, the category name is "id" and the two clusters that are created are named "Monocytes" and "Mature Ery". You can also download a full example that you can import into the AMLTutorial dataset bundled with Loupe Browser. It is a set of manual annotations for the Patient sample. You can import multiple categories at once from a single CSV file; each column will encode its own category.
With Accessibility mode selected, you can choose to import a set of feature lists or export the currently loaded set of feature lists. Click the Export button to the right of the feature list selector.
Filters can be imported and exported and used across datasets similarly to other features discussed on this page. However we recommend that you do not manually create filters. The filters use a JSON spec that is specific to Loupe Browser and is subject to modification.