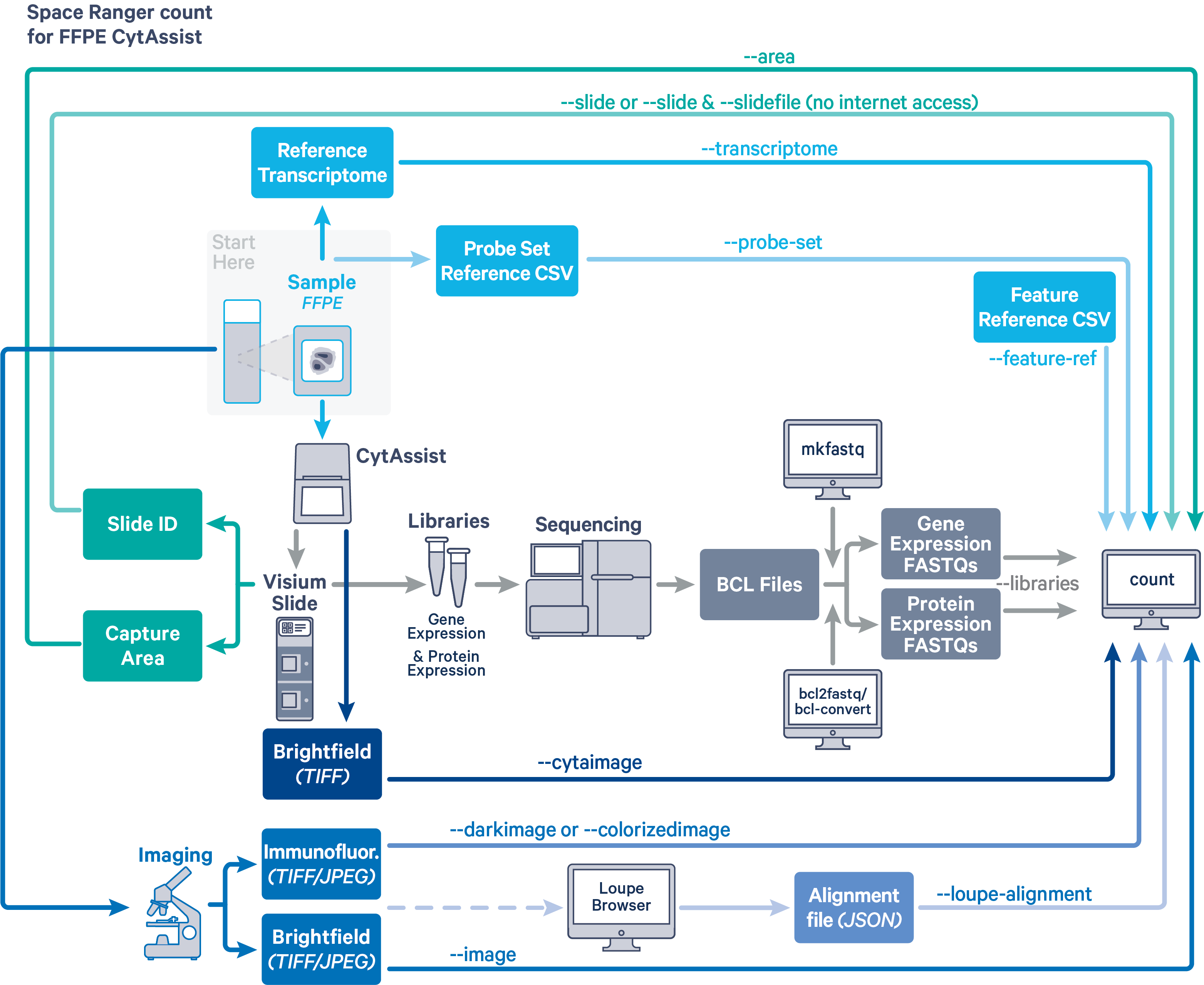
Space Ranger v2.1 and later can analyze Visium Spatial Gene and Protein Expression datasets generated using formalin fixed paraffin embedded (FFPE) samples. In this CytAssist-enabled workflow, separate gene expression (GEX) and protein expression (PEX) sequencing libraries are generated from the same Capture Area on the Visium slide. The spaceranger count pipeline is necessary to analyze these data.
Before running the pipeline, check that you have the following inputs prepared:
- Libraries CSV specifying FASTQ files for the GEX and PEX libraries (
--libraries). See the next section for details on how to set up this file. - The corresponding CytAssist image in
TIFFformat (--cytaimage) - Microscope image (optional) in either
TIFF,QPTIFForJPEGformat:--imagefor a brightfield microscope image--darkimagefor a dark background fluorescence microscope image--colorizedimagefor a composite colored fluorescence microscope image
- Slide parameters specified by:
--slide&--areaifspacerangerhas access to internet--slidefile,--slide&--areaifspacerangerhas no access to the internet. The slide layout file must be directly downloaded.--unknown-slideif Visium slide details are unknown
- The reference transcriptome (
--transcriptome). Human and mouse references are available for download, or build a custom reference. - The probe set CSV (
--probe-set). - The feature reference CSV (
--feature-ref).
The Libraries CSV (--libraries) specifies the input FASTQ data for the GEX and PEX libraries.
| Column Name | Description |
|---|---|
fastqs | Path to the directory containing the demultiplexed FASTQ files for this sample. Analogous to the --fastqs arg to spaceranger count. This field does not accept comma-delimited paths. For multiple sets of FASTQs for the same library, add an additional row, and use the use same library_type value. |
sample | Sample name assigned in the bcl2fastq sample sheet. Each unique sample ID requires a separate line in the CSV file. |
library_type | Must be Gene Expression for the GEX libraries or Antibody Capture for PEX libraries. Case-sensitive. |
The follow example includes sequencing data from two libraries that were demultiplexed into two sets of FASTQ files with prefixes GEX_sample1 (GEX) and Ab_sample1 (PEX).
| fastqs | sample | library_type |
|---|---|---|
| /path/to/GEX_FASTQs | GEX_sample1 | Gene Expression |
| /path/to/PEX_FASTQs | Ab_sample1 | Antibody Capture |
The probe set CSV files can be found in the probe_sets directory in the Space Ranger package or downloaded here.
spaceranger-2.1.0/probe_sets/
├── Visium_Human_Transcriptome_Probe_Set_v1.0_GRCh38-2020-A.csv
├── Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv
└── Visium_Mouse_Transcriptome_Probe_Set_v1.0_mm10-2020-A.csv
The example command line below is based on the public dataset Visium CytAssist Gene and Protein Expression Library of Human Tonsil with Add-on Antibodies, H&E, 6.5 mm (FFPE). This command assumes all inputs are in the current working directory; change paths as needed. A separate instance of the spaceranger count pipeline should be run on each Capture Area.
spaceranger count --id=Visium_GEX_PEX \
--probe-set=Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv \
--image=CytAssist_FFPE_Protein_Expression_Human_Tonsil_tissue_image.tif \
--cytaimage=CytAssist_FFPE_Protein_Expression_Human_Tonsil_image.tif \
--transcriptome=refdata-gex-GRCh38-2020-A \
--slide=V43J30-093 \
--area=D1 \
--libraries=libraries.csv \
--feature-ref=CytAssist_FFPE_Protein_Expression_Human_Tonsil_feature_reference.csv
By default, Space Ranger will perform automatic image registration of the two image inputs as well as fiducial alignment and tissue detection on the CytAssist image. When using the manual fiducial alignment or the manual CytAssist image alignment, specify the alignment JSON file to the --loupe-alignment option. For a complete list of arguments and options, see the Command Line Argument Reference, or run spaceranger count --help.
A successful spaceranger count run concludes with a message similar to this:
Outputs:
- Run summary HTML: /path/to/Visium_GEX_PEX/outs/web_summary.html
- Outputs of spatial pipeline:
Slide image annotated with fiducial alignment: /path/to/Visium_GEX_PEX/outs/spatial/aligned_fiducials.jpg
Slide image annotated with detected tissue: /path/to/Visium_GEX_PEX/outs/spatial/detected_tissue_image.jpg
Scale factors for hires and lowres images and fiducial and spot diameters: /path/to/Visium_GEX_PEX/outs/spatial/scalefactors_json.json
Slide image scaled to 2000px in the largest dimension: /path/to/Visium_GEX_PEX/outs/spatial/tissue_hires_image.png
Slide image scaled to 600px in the largest dimension: /path/to/Visium_GEX_PEX/outs/spatial/tissue_lowres_image.png
RGB brightfield image generated by CytAssist instrument: /path/to/Visium_GEX_PEX/outs/spatial/cytassist_image.tiff
Registration image between CytAssist image and microscope image: /path/to/Visium_GEX_PEX/outs/spatial/aligned_tissue_image.jpg
Slide position and tissue status of barcodes CSV: /path/to/Visium_GEX_PEX/outs/spatial/tissue_positions.csv
Per feature Moran's I CSV: /path/to/Visium_GEX_PEX/outs/spatial/spatial_enrichment.csv
Fluorescence intensity at spot CSV: null
- Run summary CSV: /path/to/Visium_GEX_PEX/outs/metrics_summary.csv
- BAM: /path/to/Visium_GEX_PEX/outs/possorted_genome_bam.bam
- BAM BAI index: /path/to/Visium_GEX_PEX/outs/possorted_genome_bam.bam.bai
- BAM CSI index: null
- Filtered feature-barcode matrices MEX: /path/to/Visium_GEX_PEX/outs/filtered_feature_bc_matrix
- Filtered feature-barcode matrices HDF5: /path/to/Visium_GEX_PEX/outs/filtered_feature_bc_matrix.h5
- Unfiltered feature-barcode matrices MEX: /path/to/Visium_GEX_PEX/outs/raw_feature_bc_matrix
- Unfiltered feature-barcode matrices HDF5: /path/to/Visium_GEX_PEX/outs/raw_feature_bc_matrix.h5
- Secondary analysis output CSV: /path/to/Visium_GEX_PEX/outs/analysis
- Per-molecule read information: /path/to/Visium_GEX_PEX/outs/molecule_info.h5
- Loupe Browser file: /path/to/Visium_GEX_PEX/outs/cloupe.cloupe
- Feature Reference: /path/to/Visium_GEX_PEX/outs/feature_reference.csv
- Target Panel file: null
- Probe Set file: /path/to/Visium_GEX_PEX/outs/probe_set.csv
- Normalization factors used to normalize AB counts: /path/to/Visium_GEX_PEX/outs/isotype_normalization_factors.csv
- Deconvolution directory: /path/to/Visium_GEX_PEX/outs/deconvolution
- Raw probe barcode matrix: /path/to/Visium_GEX_PEX/outs/raw_probe_bc_matrix.h5
Waiting 6 seconds for UI to do final refresh.
Pipestance completed successfully!
2023-04-08 11:47:17 Shutting down.
Saving pipestance info to "Visium_GEX_PEX/Visium_GEX_PEX.mri.tgz"
To continue, see Understanding Space Ranger Outputs. It is recommended to check the web_summary.html file first to double check the analysis was run properly and check other quality control metrics.