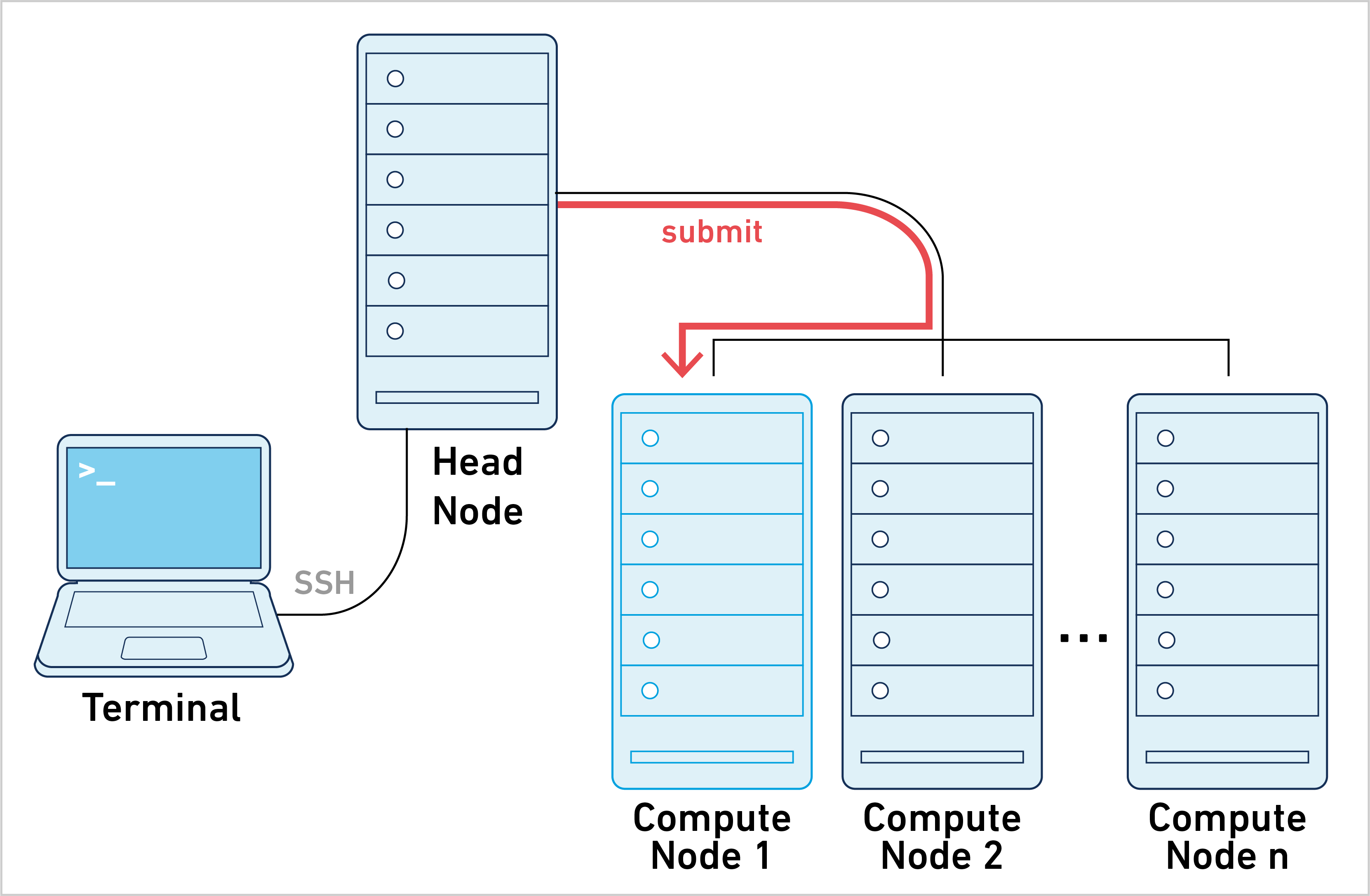
Job submission mode is one of three primary ways of running Space Ranger. To learn about the other approaches, go to the Different Ways of Running Space Ranger page.
Space Ranger can be run in job submission mode, by treating a single node from the cluster like a local server. This leverages existing institutional hardware for pipeline analysis.
Select a job scheduler below to see a sample job script.
#!/usr/bin/env bash # ================================== # Job Script # ================================== #SBATCH -J {JOB_NAME} #SBATCH --export=ALL #SBATCH --nodes=1 --ntasks-per-node={NUM_THREADS} #SBATCH --signal=2 #SBATCH --no-requeue ### Alternatively: --ntasks=1 --cpus-per-task={NUM_THREADS} # ================================== # Consult with your cluster administrators to find the combination that # works best for single-node, multi-threaded applications on your system. # ================================== #SBATCH --mem={MEM_GB}G #SBATCH -o {STDOUT_LOG} #SBATCH -e {STDERR_LOG} {PIPELINE_CMD}
#!/usr/bin/env bash #!!! This is not officially supported by 10x # ================================== # Setup Instructions # ================================== # # 1. Substitute {PE_NAME} below with name of your cluster's # shared-memory parallel environment. If your cluster does not # have a parallel environment, # delete this line. However, the job will run with only 1 thread. # ================================== # Job Script # ================================== #$ -N {JOB_NAME} #$ -V #$ -pe {PE_NAME} {NUM_THREADS} #$ -cwd #$ -l mem_free={MEM_GB}G #$ -o {STDOUT_LOG} #$ -e {STDERR_LOG} #$ -S "/usr/bin/env bash" {PIPELINE_CMD}
#!/usr/bin/env bash #!!! This is not officially supported by 10x # ================================== # Job Script # ================================== #BSUB -J {JOB_NAME} #BSUB -n {NUM_THREADS} #BSUB -o {STDOUT_LOG} #BSUB -e {STDERR_LOG} #BSUB -R "rusage[mem={MEM_MB}]" #BSUB -R span[hosts=1] {PIPELINE_CMD}
#!/usr/bin/env bash #!!! This is not officially supported by 10x # ================================== # Job Script # ================================== #PBS -N {JOB_NAME} #PBS -V #PBS -l nodes=1:ppn={NUM_THREADS} #PBS -l mem={MEM_GB}gb #PBS -o {STDOUT_LOG} #PBS -e {STDERR_LOG} {PIPELINE_CMD}
Copy the sample job script to a text editor, and replace all the variables enclosed within braces (for example, {JOB_NAME}or {STDOUT_LOG}) with literal names, paths, and values.
The last line, {PIPELINE_CMD} , is replaced by a Space Ranger command, specifying --jobmode=local, --localcores={NUM_THREADS} and --localmem=0.9*{MEM_GB} to restrict CPU and memory use that's consistent with the directives in the job script.
For example, if the job script requests 8 cores and 64 GB of memory from the cluster, then the Space Ranger command should include --jobmode=local --localcores=8 --localmem=57.
If the sample job script is saved as runJob.bash, then following command submits the job to the cluster for each corresponding job scheduler.
sbatch runJob.bash
qsub runJob.bash
bsub runJob.bash
qsub runJob.bash