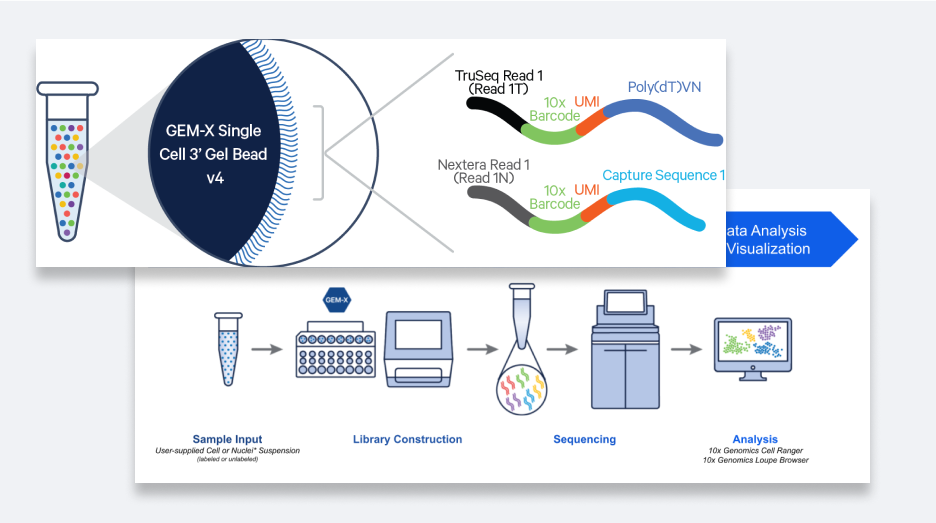
Chromium单细胞分析
为您的成功而打造的单细胞分析
单细胞分析能够带来有意义的发现
批量细胞测序产生了平均读数,遗漏了驱动复杂生物学的关键细节。 单细胞测序让科学家们能够看到每个细胞的独特基因表达模式。这种视野能够更全面地表征组织异质性,揭示对健康和疾病有重大影响的稀有细胞类型。
Chromium单细胞分析的优势
更高的基因检测灵敏度,细胞回收率高达80%。
增强对具有挑战性的细胞类型的检测,包括中性粒细胞。
通过最关键步骤的自动化,减少手动操作时间,降低出错风险。
生成更高质量的文库,能够以较低深度进行测序。
以10年经验、2,200多项专利和超过15亿美元的研发投资作为后盾。
创新,为您每一步的成功而开发
分析更多样本类型
利用性能可靠的优化方案分析新鲜、冷冻或固定样本,甚至是FFPE样本
关键步骤自动化,减少出错
在短短几分钟内生成多达200万个带有条形码的微滴,细胞回收率高达80%
构建高质量的测序级文库
生成可用读数高达95%的文库,让您能以更低的测序成本检测更多基因
轻松分析和发现
利用功能强大的软件工具对数据进行处理和可视化,无需生物信息学经验

与生俱来的性能、重现性和经济性
广泛而灵活的分析组合
Flex基因表达
按照您的日程表进行固定、分批和运行。
基于探针的化学方法: 在低质量样本和FFPE样本上表现出色
同一个细胞的多组学读数: 基因表达,蛋白质,CRISPR
高通量的选择: 每次运行可分析多达200万个细胞(1-16个样本)
通用型基因表达
不限物种,适用性最广。
基于逆转录的化学方法: 提供最广泛的信息,包括异构体、SNP等
同一个细胞的多组学读数: 3’或5’基因表达,TCR/BCR,蛋白质,CRISPR
用于具有成本效益的研究: 每次运行最多 160,000 个细胞(1-8 个样本
表观基因组和染色质分析
揭开表观基因组图谱。
ATAC-seq化学方法: 探索开放染色质区域,与3’基因表达直接关联(多组学试剂盒)
同一个细胞的多组学读数: 染色质可及性,3’基因表达
可扩展的细胞核制备: 每次运行可分析多达80,000个细胞核(1-8个样本)
在过去几年里,我们注意到单细胞技术取得了很多进展,这要归功于10x Genomics。这种多组学方法对科学家和医生了解人类疾病特别有用。David Michonneau博士法国圣路易医院血液学教授
提升您的研究成果
将Chromium带入您的实验室
| 平台 | |||
| 何时使用 | Comprehensive single cell data适用于细胞群体和状态的深度表征 | High-resolution spatial gene expression了解复杂组织、邻域和细胞间相互作用。与其他空间多组学、组织学和形态学方法整合。 | |
| 为何使用 | 无偏的单细胞发现
每个基因的灵敏度高 | 无偏的空间发现 | 靶向的空间探索
每个基因的灵敏度高 |
| 应用 | 全转录组基因表达
蛋白质
TCR、BCR
CRISPR
ATAC | 全转录组基因表达 | 靶向基因表达(多达5000个基因) |
| 分辨率 | 单细胞 | 分配在2-µm区域内的转录本 | 单细胞 |
| 数据读出 | 基于NGS | 基于NGS | 基于成像 |
| 样本兼容性 | 来自新鲜、冷冻或FFPE样本的单细胞或单细胞核悬液 | FFPE样本 | 新鲜冷冻样本
FFPE样本 |