Choose a product below to filter the page content to your needs:
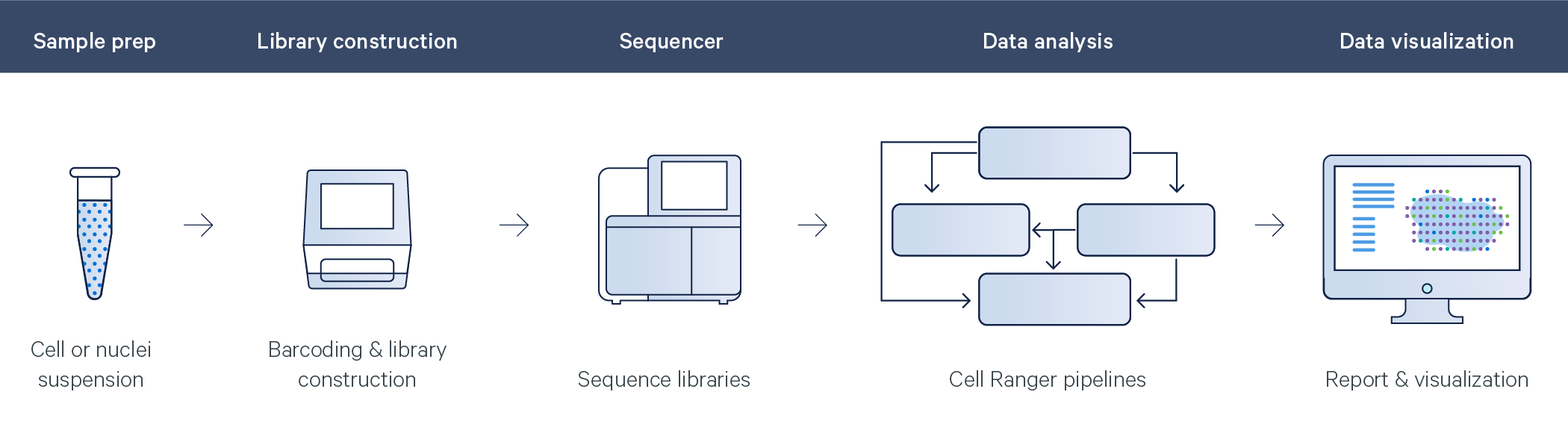
Cell Ranger processes all Feature Barcode data through a counting pipeline called cellranger count that quantifies each feature in each cell. The pipeline outputs a unified feature-barcode matrix that contains gene expression counts alongside Feature Barcode counts for each cell barcode. The feature-barcode matrix replaces the gene-barcode matrix emitted by older versions of Cell Ranger.
The pipeline first extracts and corrects the cell barcode and UMI from the feature library using the same methods as gene expression read processing. It then matches the Feature Barcode read against the list of features declared in the Feature Barcode Reference. The counts for each feature are available in the feature-barcode matrix output files and in the Loupe Browser output file.
To enable Feature Barcode analysis, cellranger count needs two new inputs:
- Libraries CSV is passed to cellranger count with the --libraries flag, and declares the FASTQ files and library type for each input dataset. In a typical Feature Barcode analysis there will be two input libraries: one for the normal single-cell gene expression reads, and one for the Feature Barcode reads. This argument replaces the --fastqs argument.
- Feature Reference CSV is passed to cellranger count with the --feature-ref flag and declares the set of Feature Barcode reagents in use in the experiment. For each unique Feature Barcode used, this file declares a feature name and identifier, the unique Feature Barcode sequence associated with this reagent, and a pattern indicating how to extract the Feature Barcode sequence from the read sequence. See Feature Barcode Reference for details on how to construct the feature reference.
When inputting Feature Barcode data to Cell Ranger via the Libraries CSV file, you must declare the library_type of each library. Specific values for library_type will enable additional downstream processing, specifically for Antibody Capture, Antigen Specificity, or CRISPR Guide Capture. The following table outlines the types of libraries that can be specified and what they mean for the downstream processing.
code block
| 10x Solution | Gene Expression | Cell Surface Protein |
|---|---|---|
| Single Cell Gene Expression 3' v2 | - | |
| Single Cell Gene Expression 3' v3 | ||
| Single Cell Gene Expression 3' v3.1 | ||
| Single Cell Gene Expression 3' v3.1 (Dual Index) | ||
| Single Cell Gene Expression 3' HT v3.1 (Dual Index) | ||
| Single Cell Gene Expression 3' LT v3.1 (Dual Index) |