Bulk vs. single cell RNA-seq: Experimental differences and advantages of single cell resolution
Single cell sequencing allows scientists to define cell type–specific gene expression and multiomic readouts, resolving the cellular heterogeneity that drives the expression patterns seen in bulk RNA-seq.
Keep reading to learn about these two approaches to studying gene expression, bulk RNA sequencing and single cell RNA sequencing, their respective advantages and disadvantages, and how single cell RNA sequencing has impacted crucial areas of research to enable breakthroughs in our knowledge of biological complexity and disease.
Jump ahead to:
- Different approaches to gene expression: Bulk vs. single cell RNA-seq
- Impact of single cell RNA-seq and future trends
- Getting started with single cell RNA-seq
Different approaches to studying gene expression: Bulk vs. single cell RNA-seq
There are two major approaches to detect gene expression levels from cells in a complex biological sample that we’ll address in this blog: bulk RNA sequencing (bulk RNA-seq) and single cell RNA sequencing (scRNA-seq). To understand the strengths and limitations of each approach, let’s first establish what they are and how they’re different.
What is bulk RNA-seq?
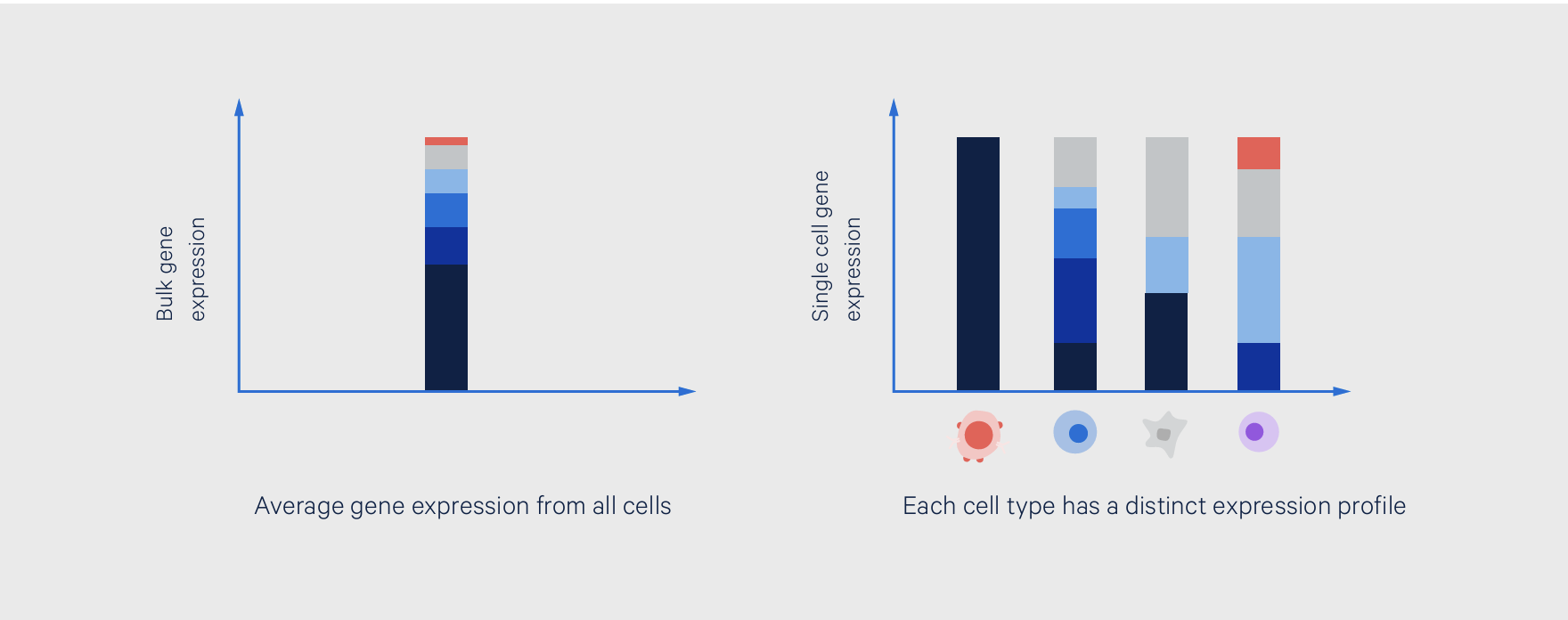
Bulk RNA-seq is a next-generation sequencing (NGS)-based method to measure the whole transcriptome across a population of cells. It is a whole population approach, meaning it provides a readout of the gene expression profile for the entire sample, with many different cells pooled together to contribute to this profile. It is also an average readout, meaning it provides the average expression levels for individual genes across all cells that compose the sample.
Steps in a bulk experiment
In the bulk RNA-seq workflow, your biological sample is digested to extract RNA. This could be total RNA from the sample, or steps could be taken to enrich mRNA or deplete ribosomal RNA. The RNA is converted into cDNA and followed by more processing steps to prepare a sequencing-ready gene expression library. After sequencing and data analysis, you can review gene expression levels across the tissue sample.
What is single cell RNA-seq?
Single cell RNA sequencing (scRNA-seq) is a method for studying the whole transcriptome gene expression profile of each individual cell from a sample. While a bulk readout could be compared to the view of a forest, a single cell readout is like seeing every tree. As such, there are a few key experimental differences, which we cover below.
Sample preparation for single cell RNA-seq
scRNA-seq measurements come from individual cells, as opposed to a pool of cells. First, you need to generate viable single cell suspensions from whole samples that have been digested through an enzymatic or mechanical process, cell sorting, or other cell isolation techniques. This is followed by cell counting and quality control steps to ensure samples have an appropriate concentration of viable cells and are free of clumps and cell debris.
If desired, you can also stain your sample with antibodies to label proteins and other biological analytes, or perform FACS enrichment for cell types of interest.
The steps taken to dissociate a sample will vary based on your starting material and the goals of the experiment. Additional preparation steps may be necessary depending on the quality of the tissue, sample abundance, cell size, or the need to extract nuclei for chromatin accessibility profiling. Regardless of these specific considerations, the same basic principle of generating a high-quality single cell suspension applies across sample types.
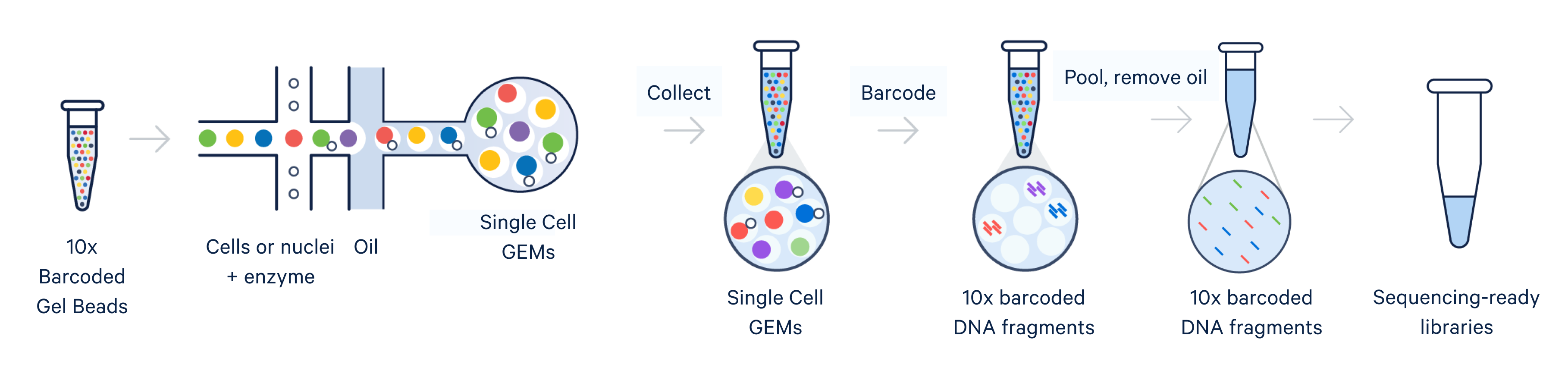
Instrument-enabled cell partitioning
Single cell RNA-seq, like bulk RNA-seq, has the goal of isolating RNA and converting it to cDNA, enabling measurement of gene expression across the whole transcriptome through next-generation sequencing. For the 10x Genomics scRNA-seq workflow, single cells are isolated into individual micro-reaction vessels (Gel Beads-in-emulsion, or GEMs) before the RNA is isolated. This partitioning step happens in an automated, controlled environment within a microfluidic chip loaded onto a Chromium X series instrument.
Subsequently, within the GEM, Gel Beads are dissolved, releasing oligos containing unique barcodes. Additionally, the cell is lysed, allowing RNA to be captured and barcoded with cell-specific barcodes. This ensures analytes from each cell can be traced back to their cell of origin. Barcoded products are then used to create a sequencing library, allowing measurement of gene expression across the entire transcriptome of each individual cell.
Applications of bulk RNA-seq
Since bulk readouts provide a holistic view of the average gene expression profile of the sample, some key use cases (1) for this approach include:
-
Differential gene expression analysis: By comparing bulk gene expression profiles between different experimental conditions, including disease vs. healthy, treated vs. control, developmental stages, or time-course experiments, you can identify the distinct genes that are upregulated or downregulated in these conditions. This can also support applications like:
- Discovery of RNA-based biomarkers and molecular signatures for diagnosis, prognosis, or stratification of diseases
- Investigating how sets of genes (pathways and networks) change collectively under various biological conditions
-
Tissue or population-level transcriptomics: Bulk data can also help you obtain a global expression profile from whole tissues, organs, or bulk-sorted cell populations. This population-level analysis can be useful for:
- Large cohort studies or biobank projects
- Providing baseline transcriptomic profiles for new or understudied organisms or tissues
- Supporting deconvolution studies with single cell RNA-sequencing reference maps
-
Identifying and characterizing novel transcripts: Bulk data can be used to annotate isoforms, non-coding RNAs, alternative splicing events, and gene fusions.
Applications of single cell RNA-seq
With a single cell gene expression readout, you can resolve more layers of biological complexity within heterogeneous samples, including teasing out rare or low-abundance cell types and cell states, and revealing mechanisms underlying cell function and differentiation.
Some of the key use cases of a single cell approach, and the questions that you can answer with it, include:
Characterizing heterogeneous cell populations, including novel cell types, cell states, and rare cell types
- What cell types or states are present in a tissue (e.g., neurons vs. astrocytes in brain tissue)?
- What are the proportions of different cell types or states?
- What are the gene expression differences between similar cell types or subpopulations (e.g., different subtypes of T cells)?
- How do gene expression programs vary within a supposedly homogeneous cell type (e.g., cycling vs. quiescent states, activated vs. resting immune cells)?
- Are there rare cell types or transient states that play key roles in biology?
Discovering new cell markers and regulatory pathways
- What are the co-expression patterns of genes at the single cell level?
- How does knocking out certain genes affect other gene programs in unique cell populations?
Reconstructing developmental hierarchies and lineage relationships
- How does cellular heterogeneity evolve over time (e.g., during development or disease progression)?
Profiling healthy and diseased tissue, organs, and systems
- How do individual cells respond to stimuli or perturbations, such as treatment or disease conditions?
- Are certain cells or cell states the major drivers of disease biology or treatment resistance?

Bulk and single cell work together to reveal a new druggable target in B-ALL
In their 2024 Cancer Cell paper, Huang et al. leveraged both bulk and single cell RNA-seq in healthy human B cells and leukemia clinical samples to identify development states driving resistance and sensitivity to a common chemotherapeutic agent, asparaginase, against B-cell acute lymphoblastic leukemia (B-ALL). See what they discovered.
Advantages and disadvantages of bulk RNA-seq
The bulk RNA-seq workflow has the advantage of being a lower-cost option for whole transcriptome analysis with a focus on population-level insights. The lower sequencing depth requirements further reduce costs. Moreover, simpler requirements for sample prep and lower data complexity and resolution make it an easier workflow with more straightforward analyses.
However, these advantages come with major tradeoffs in resolution and the potential for biological discovery. Because bulk RNA-seq provides an average readout of the gene expression across all the cells in a sample, it is unable to tease apart the cellular origins of gene expression readouts. This would mask if one or a few cell types are the majority producers of a certain gene or unique transcript.
This also means that a bulk readout cannot reveal the cellular heterogeneity of a sample, making it less ideal for highly heterogeneous tissues. In contrast, single cell sequencing can reveal this heterogeneity, even bringing rare, low-abundance cell types that could be driving unique gene expression profiles into view.
Advantages and disadvantages of single cell RNA-seq
Single cell RNA-seq presents the powerful advantage of offering whole transcriptome profiling at the resolution of individual cells, which advances biological discovery like never before. The single cell technology that 10x Genomics provides comes with the added benefits of a robust, reproducible, instrument-enabled workflow that reduces error by automating critical steps, ensuring the highest data quality.
However, single cell workflows come with the perceived tradeoffs of cost and complexity. On the pricing side, single cell RNA-seq is typically associated with a higher cost versus bulk measurements. This is, in part, because deeper sequencing is often required to capture rare features, increasing overall experiment costs.
In terms of complexity, single cell RNA-seq requires sample dissociation into a single cell suspension and high cell viability, making for a more technically complex sample prep workflow. Furthermore, unfamiliar analysis may be necessary to unlock the full potential of single cell datasets.
But hard things are often the most rewarding. The value that single cell analysis can bring to your research, as you can see in the discoveries it has enabled, is well worth any friction you may experience as you try to learn how to perform this technique.
More importantly, many of the perceived challenges associated with single cell RNA sequencing are actually being continuously addressed and alleviated, making this technique more accessible than ever.
Cost
Cost is often a major barrier for new users—especially those who use bulk. But the low per-sample cost of bulk analysis doesn't always translate to long-term savings. Without high resolution, you could miss a new cell type or a low-level transcript key to moving your project forward (or publishing in a high-impact journal).
With 10x Genomics, you not only get single cell resolution, but also higher gene sensitivity, fewer errors, and the confidence to get it right the first time. And we’re taking huge steps to make Chromium Single Cell work with your budget:
- The GEM-X Flex Gene Expression assay is bringing down cost per cell by enabling high-throughput single cell experiments. Plus, Flex maximizes your sequencing budget with highly sensitive transcript detection.
- GEM-X Universal 3’ and 5’ Multiplex assays are lowering per-sample costs with smaller kit options that let you input smaller amounts of your sample and use fewer reagents.
- The Chromium Xo instrument offers an affordable entry point to high-performance single cell profiling with our Universal 3' Gene Expression assay.

Choosing the right single cell assay for your experiments
Explore your options for single cell research in our blog, offering a breakdown of the single cell RNA-seq assay families in the Chromium platform, Flex and Universal. See their advantages, plus practical considerations for experiment setup, to find guidance on choosing the right single cell assay.
Experimental difficulty
New sample prep requirements and an unfamiliar instrument-based workflow may seem daunting, but, in reality, every single cell experiment follows three common steps that are fundamentally similar to a bulk experiment: sample prep, running the assay, and data analysis.
We have sample prep methods optimized for your success, including 40+ Demonstrated Protocols across our Chromium Single Cell portfolio, plus helpful explainer videos and User Guides. Our support resources also include expert guidance and coaching through sample preparation and experiment design from a global team of Field Application Scientists.
Moreover, the push-button simplicity of the Chromium X instrument series instills confidence, offering superior reproducibility and time savings. We automate the most critical step of any single cell workflow, partitioning thousands of cells in minutes with just 3–4 hours of hands-on time, minimizing errors from manual workflow pipetting. And if you aren’t ready to own an instrument, you can work with one of the many core facilities that offer Chromium Single Cell services.

The results of this robust, instrument-enabled partitioning speak for themselves: you can get more from every sample with up to 80% cell recovery, including low cell inputs, and capture of fragile cells. And you can ensure consistent performance with an R² of 0.97 across runs, users, and timepoints.
Learn more about Chromium Single Cell workflows, built for your success.
Data analysis
We want to do everything in our power to make single cell data accessible and interpretable for you, no bioinformatics experience needed. Our free, user-friendly software was developed to accelerate your access to experimental conclusions:
- Cell Ranger analysis pipelines process scRNA-seq data, providing results quickly and with generous free limits on 10x Genomics Cloud Analysis, our optimized cloud infrastructure.
- Visualize your processed data with the interactive Loupe Browser software, quickly plotting differential expression of your favorite genes.
- Take advantage of continued innovations that are further streamlining data analysis, including automated cell annotation (in beta).
Using these tools, you can identify cell types and states, detect rare transcripts, perform multi-sample comparisons and more. And you’re not alone in getting started. Our Analysis Guides offer a collection of introductions, tutorials, and blogs for single cell data analysis that are helpful whether you’re a seasoned pro or looking at your first dataset.
Impact of single cell RNA-seq and future trends
While bulk RNA-seq has powerful applications, there are some things it can’t do. Some discoveries can only happen through the application of single cell technology. There are thousands of publications demonstrating the utility and impact of single cell RNA-seq, but we wanted to highlight a few examples:
Finding the cellular source of the cystic fibrosis mutation
The discovery of the "pulmonary ionocyte," a cell type that makes up a mere 0.5% of all lung cells yet is the main source of the mutated CFTR gene causing cystic fibrosis pathology, demonstrates how identifying new cell types with single cell sequencing technology can be extremely powerful for disease research (2).
Identifying immune-checkpoint resistant cell states in melanoma
In their search for more durable therapeutic approaches for individuals with melanoma, researchers leveraged single cell RNA sequencing to reveal malignant cell states associated with immune checkpoint inhibitor resistance (3).
Revealing cellular programs underlying severe COVID-19 pathology
A single cell atlas study of severe COVID-19 autopsy tissue revealed that myeloid and epithelial cells were the primary cell types enriched for SARS-CoV-2 RNA and pointed to a cellular mechanism for lung failure (4).

Future trends: Spotlight on Billion Cells Project
The Chan Zuckerberg Initiative (CZI) recently launched the Billion Cells Project to generate an unprecedented one billion cell dataset to fuel rapid progress in AI model development in biology. Learn how our single cell technology delivers the quality and scale needed to build trustworthy models that can benefit many domains of biological research.
Getting started with single cell RNA-seq
Bulk and single cell RNA-seq are all at once similar, different, and complementary. Both methods seek to understand the transcriptome with an NGS-based readout, but at different resolutions: tissue-wide vs. single cell.
Single cell technology offers the fundamental advantage of resolving the biological heterogeneity that remains hidden in a bulk readout. From identifying novel cell types and states to revealing the underlying mechanisms of disease, therapeutic response, and resistance, insights enabled by single cell sequencing are fueling groundbreaking research with the potential to transform biology and medicine.
So what’s the next step for you and your research projects?
Continue exploring the Chromium Single Cell platform with our beginner’s guide to single cell RNA sequencing. Or, speak with a sales specialist to find the right single cell assay for your experimental goals.

Get started with scRNA-seq
Speak to your sales representative to access expert guidance on choosing the right single cell assay or deciding on an experiment design that is customized to your budget. We’re here to help you accomplish your research goals!
References:
- https://sampled.com/bulk-rna-sequencing-vs-single-cell-rna-sequencing/
- Montoro D, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560: 319–324 (2018). doi: 10.1038/s41586-018-0393-7
- Jerby-Arnon L, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175: 984–997.e24 (2018). doi: 10.1016/j.cell.2018.09.006
- Delorey TM, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 595: 107–113 (2021). doi: 10.1038/s41586-021-03570-8
About the author:

Blog contributors:




