Introducing BEAM (Barcode Enabled Antigen Mapping): Benefits of rapid, antigen-specific B- and T-cell discovery
Right now, there are billions of B and T cells circulating in your blood and crowding the halls of your bone marrow, spleen, and thymus. Some estimates put the total count at as many as 10 billion B cells and 25 million to 1 billion T cells (1,2).
But finding antigen-specific B and T cells is like finding a needle—or a breakthrough immunotherapy—in a haystack. Currently, many researchers slog through experiments for weeks or months to get a few antigen-specific hits that too often fail to reveal rare, therapeutically desirable clonotypes.
BEAM (Barcode Enabled Antigen Mapping), our newly developed antigen-specific B- and T-cell discovery workflow, excels where conventional methods have failed.
BEAM lets scientists capture high-resolution cellular profiles of antigen-specific B and T cells with a multiplexed antigen screening workflow built on our Chromium Single Cell Immune Profiling solution. In a mere week’s time, researchers can generate tens to hundreds of high-quality antigen-specific hits, offering unparalleled speed and cellular characterization. Now, both academic and translational researchers have an end-to-end solution to unearth potent antibodies, reveal novel cancer immunotherapies, and inform vaccine development for patients who don’t have time to wait.
Read on to dive deep into the challenges of antigen-specific B- and T-cell discovery, the science behind BEAM, and how you can use it to make the next big therapeutic breakthrough.
[Want to skip ahead? Use the links above to jump to sections within the article.]
Unlocking B- and T-cell therapeutic potential
B and T cells perform a vital function in the adaptive immune system, recognizing molecules the body has determined are foreign, called antigens, and subsequently triggering events that will clear those molecules and the pathogens they’re derived from, such as viruses or bacteria. This function is enacted through antigen recognition by B- and T-cell receptors (BCRs/TCRs).
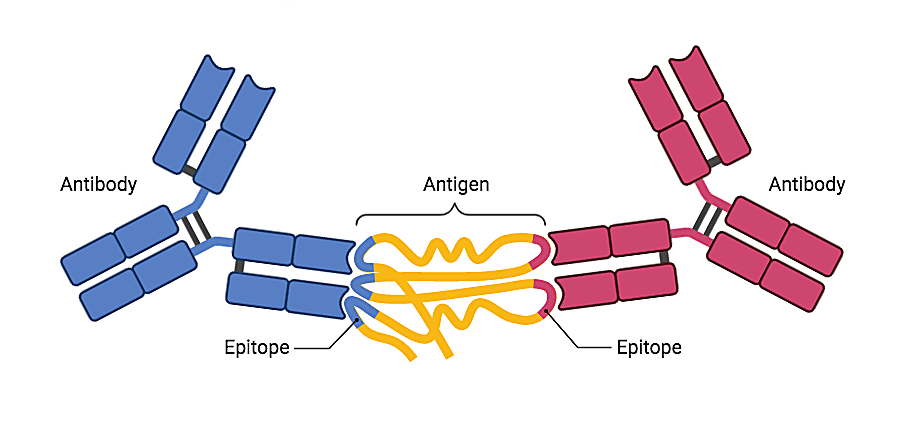
The full diversity of the existing pool of BCRs/TCRs in the body is known as the functional adaptive immune repertoire. And the key there is functional. As receptors engage with antigens, B and T cells are drafted to perform their immune functions. Antigen-reactive antibodies are unleashed to neutralize pathogens, killer T cells set out to destroy diseased cells—or, unfortunately, in the case of dysfunction, B and T cells that recognize self-antigens drive the deleterious effects of autoimmunity.
BCRs and TCRs have variable antigen binding regions, encoded by V(D)J gene sequences that can be rearranged or mutated, creating staggering permutations of possible antigen-reactive receptors. Calculating the number of possible unique V(D)J combinations reveals that, in theory, there are 1018 possible BCRs (3) and 1015 possible TCRs (4).
The adaptive immune system can generate more B- and T-cell clonotypes than the body can handle—1015 individual T cells would weigh about 500 kilograms, or a little over 1,100 pounds (4). These seemingly infinite receptors could bind to and support the clearance of a huge variety of antigens and transform therapeutic research.
Researchers have already discovered receptor–antigen relationships that translated to powerful therapies. The anti-PD1 monoclonal antibody, which binds to PD-1 receptors on T cells to block immune inhibitory signaling and activate T-cell anti-tumor function, is a highly effective therapy for a variety of cancers (5). CAR T-cell therapies are also used to treat some cancers and show promise for treating autoimmune diseases, such as lupus (6).
But these treatments only work in a subset of patients. Moreover, outside of cancer, autoimmunity, and infectious disease, we are still learning how many diseases are actually mediated through or influenced by immune cells—including neurodegenerative disorders (7). If researchers can tap into the functional adaptive immune repertoire, they will unleash unprecedented therapeutic potential.
Common hurdles to antigen-specific B- and T-cell discovery
So, how can we begin to systematically understand and leverage the functional adaptive immune repertoire to address fundamental problems in disease and medicine? It’s not so easy.
Matching antigens to their corresponding BCRs/TCRs has been historically difficult. Common methods for antibody discovery such as hybridoma technology and phage display are accompanied by weeks to months of experimental struggle. Hybridoma-based methods often only yield 1–3 expanded clones against just one antigen that require 8–12 weeks of further validation—a whopping total of 6–9 months of experimentation (8).
These methods also limit what samples researchers can use. For example, hybridomas are the product of a fusion of myeloma cells and splenocytes (8), but therapeutically desirable BCRs could be found in blood samples, bone marrow, lymph node aspirates, and more.
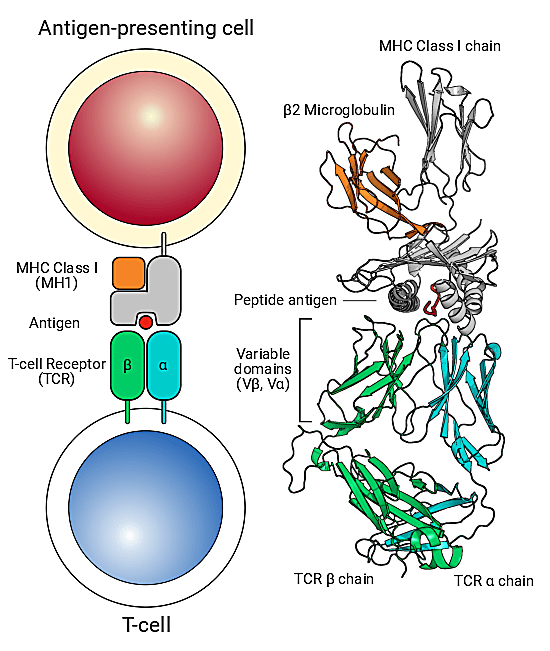
Antigen-specific T-cell discovery is no less challenging largely due to the finicky nature of TCR binding and practical considerations for cellular isolation. T cells have very specific binding requirements: they only like binding to an antigen when it’s displayed in the perfect holding device, called the major histocompatibility complex (MHC), which is displayed on the surface of antigen-presenting cells. However, it’s hard to develop stable peptide–MHC complexes to tag and subsequently isolate antigen-specific T cells from a sample—MHC molecules are large and tend to collapse, making homebrew stabilization attempts unreliable and painstaking.
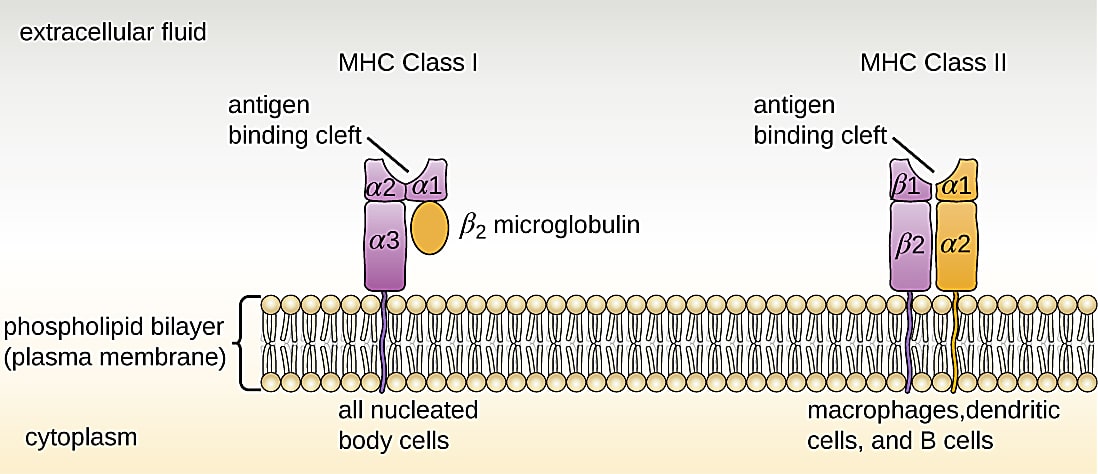
T cells with therapeutically relevant antigen specificities tend to occupy a small proportion of the vastly diverse T-cell landscape. Researchers often use fluorescence-based sorting approaches to identify and collect antigen-specific T cells from a sample. But this approach is far from perfect. Researchers can miss rare antigenic T-cell populations due to the limited number of unique fluorophores to label peptide–MHC molecules. Additionally, current methods only provide a readout of the number of antigen-specific T cells in a sample and therefore offer limited insights into T-cell states or function and the larger immune response.
Researchers have leveraged conventional methods of antibody discovery and antigen-specific T-cell isolation to make crucial immunotherapeutic discoveries, but the process is still like searching for a needle in a haystack. Researchers need faster, more robust methods to characterize immune cell phenotypes and antigen specificity and resolve rare clonotypes, not experiments that yield few antigen-specific hits from limited compatible samples.
How BEAM helps to identify antigen-specific clonotypes
We developed BEAM (Barcode Enabled Antigen Mapping) to overcome the technical challenges that delay current antigen-specific B- and T-cell discovery workflows and accelerate both academic research and therapeutic applications.
BEAM is a multiplexed antigen screening workflow that empowers rapid discovery of antigen-specific B-cell (BEAM-Ab) and T-cell (BEAM-T) clonotypes. Built on the proven Chromium Single Cell Immune Profiling solution, it uniquely allows researchers to obtain comprehensive antigen-specific cellular profiles, including full-length paired V(D)J sequences, gene expression, and cell surface proteins from the same single cell.
The BEAM workflow starts with your antigens of interest: researchers can screen binding specificities of up to 15 antigens and a control against hundreds of thousands of B or T cells in a single, one-week experiment. This means you could generate tens to hundreds of antigen-specific hits from the same sample—including compatible samples, such as PBMCs, splenocytes, lymph node aspirates, and enriched B or T cells.
Isolating antigen-specific B and T cells out of a complex sample just got easier too. Researchers can confidently identify rare clonotypes and minimize cell loss since the antigens are coupled to a uniquely barcoded BEAM Conjugate complex that contains a PE fluorescence marker. Flow sorting steps to isolate stained antigen-specific clonotypes simply come down to a check for fluorescence, rather than multicolor analysis with limited parameters.
Having a stable peptide–MHC complex isn’t a problem either. BEAM-T reagent assemblies are powered by kitted, custom loadable MHC monomers, allowing researchers to simply load their antigenic peptide of interest into an empty monomer, then stain T cells. This provides flexibility to design and source peptides from any vendor.
The BEAM workflow also includes easy-to-use data analysis and visualization software to explore and interpret antigen–clonotype relationships. Antigen specificity scores are calculated to identify antigen-specific clonotypes by comparing UMI counts associated with antigens of interest against UMI counts associated with the control antigen.
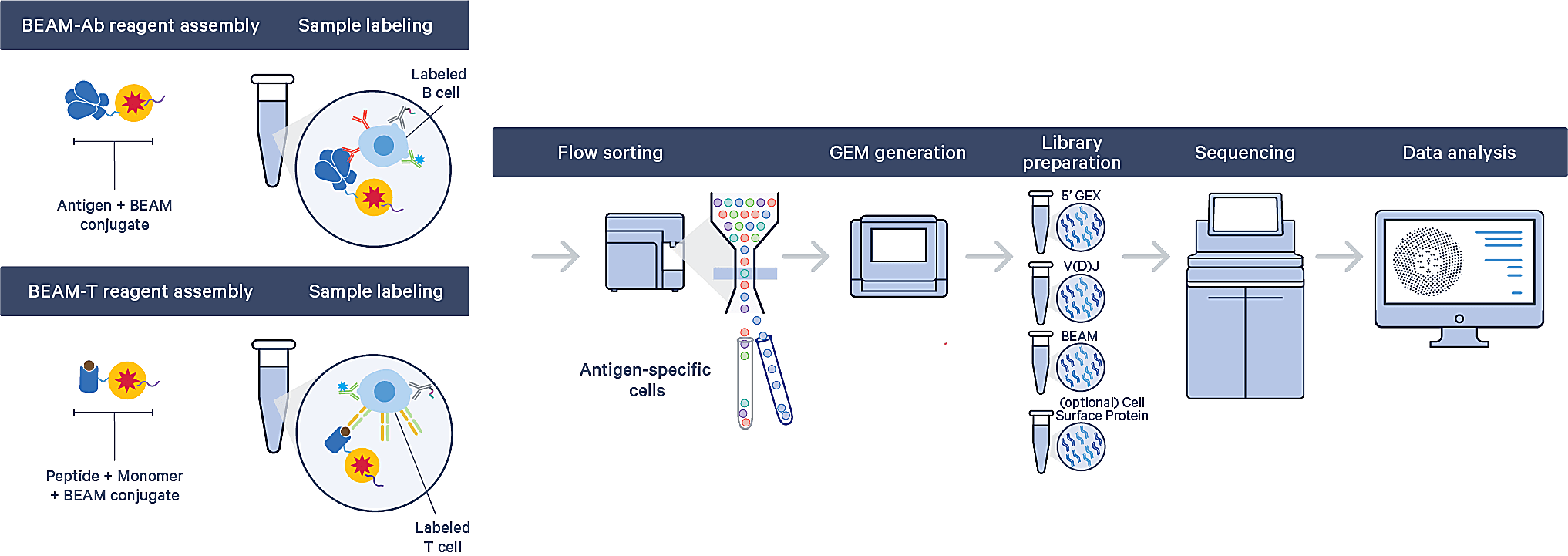
In the following sections, we’ll dive into how BEAM streamlines antigen-specific clonotype discovery experiments and how it can be applied to real-world research examples—from identifying TCRs specific to cancer neoantigens, to surveying and cataloging rare antigen-specificities within the post-COVID immune landscape.
Some of the ways BEAM will advance science
How can multiplexing and screening up to 15 antigens in a single experiment help your research? What is the benefit of capturing a comprehensive cellular profile, including gene expression and full-length, paired V(D)J sequences, alongside antigen specificity? We have some ideas.
Antibody discovery
Let’s say you’re an infectious disease researcher or a clinical scientist working on targeted immunotherapies. You’re on the hunt for potent antibodies against a known antigen target, such as a viral peptide or a surface receptor on cancer cells. These antigen targets have different epitopes that different antibodies bind to with varying affinity. Further, they often mutate or recombine as viruses or cancer cells evolve, which can alter antibody affinity as well. Screening B cells against a more comprehensive antigen panel increases your chances of finding the most potent neutralizing antibodies.
In a recent internal study, 10x Genomics scientists validated this approach using BEAM-Ab to identify antigen-specific immune cells against the SARS-CoV-2 spike protein, summarized in this poster. They developed a panel of three antigens that included pre-fusion trimeric spike proteins, such as trimeric spike antigen S and trimeric D614G, and a human control protein, then assayed PBMCs from a convalescent COVID-19 survivor. In a one-week workflow, they identified hundreds of antigen-specific B-cell clonotypes. They also obtained full-length, paired V(D)J sequences for specific BCRs, which can be used to fast-track production of recombinant antibodies for downstream validation. We expect this workflow will enable rapid, robust antibody discovery beyond our study.
Cancer immunotherapeutics
We’re just beginning to understand how powerful immunotherapies can be against cancer, and BEAM promises to accelerate crucial translational oncology research. Let’s say you’re interested in identifying patient T cells specific to cancer neoantigens—antigens derived from a patient’s cancer cells that the immune system could recognize as foreign and subsequently target. There could be dozens of candidates for the ideal neoantigen target from a single patient, suggesting the value of multiplexed antigen screening by BEAM-T to reveal the full breadth of antigen-specific T-cell clonotypes in the patient sample.
Researchers can use the unparalleled cellular resolution provided by BEAM-T to evaluate T-cell phenotypes, better understand how antigen–receptor interactions influence effector functions, and ultimately guide development of personalized cancer vaccines. And access to full-length, paired TCR sequences forges a path to investigate the potential of receptors for adoptive cell therapy, such as CAR T-cell therapy.
Vaccine development
In the infectious disease arms race, we need all the tools that we can get to better understand the cellular and molecular immune response to pathogens as well as to vaccines. Let’s say you’re trying to understand which antigenic peptides produce the strongest antiviral immune response in the body to inform vaccine development. With BEAM-T, you could screen against multiple viral peptides, including antigens from different strains of the same virus, in one experiment to track their effects on the T-cell immune response. You could identify which antigens drive a unique immune response and, ultimately, inform development of a multivalent vaccine that ensures the broadest protection against all viral strains.
“BEAM technology will greatly advance our understanding of infectious disease biology and will speed the discovery of exceptionally rare monoclonal antibodies with unique, therapeutically desirable specificities,” said Bryan Briney, PhD, an assistant Professor at Scripps Research Institute. “The BEAM workflow generates rich and powerful data that helps our understanding of variant-resistant immunity and will play a key role in the design and development of durable vaccines.”
From receptor databases to functional knowledge
A number of immune consortia—such as the Immunological Genome project—recently formed to build out large repositories of BCR/TCR repertoire data and, in turn, establish standard processes for the public sharing of these large datasets in order to better utilize repertoire data to understand immune-mediated diseases and therapies.
BEAM is uniquely positioned to help scale these cataloging efforts like never before, offering rapid insights into a previously unobtainable diversity of antigen-specific BCR/TCR repertoire data. This data could serve as the springboard for faster vaccine development, tracking the immune response in autoimmune and infectious disease, and identifying novel therapeutics against cancer.
BEAM—shedding light on immune potential
BEAM is a transformative approach to the search for therapeutically relevant antigen-specific B- and T-cell clonotypes, streamlining a process typically riddled with challenges and delays. But the true readout of its value will come through the innovation and ingenuity of the scientists who set new ideas and applications in motion to reach their moonshots—novel therapies and cures.
BEAM peels back just one layer of immune complexity, a complexity that extends from single cells to tissues and beyond. Improved access to antigen-specific clonotypes begs new questions. Which immune cells can penetrate the tumor microenvironment? How can you begin to trace and understand the function of a therapeutically relevant TCR in the context of actual tissue?
These are the questions that we at 10x Genomics look forward to answering, together with you, the scientists who inspire and challenge us daily to push the boundaries of what is possible.
Learn more about BEAM by exploring resources on our application page →
References:
- Dr. Biology. "B-cells". ASU - Ask A Biologist. 16 February, 2011. https://askabiologist.asu.edu/b-cell
- Dr. Biology. "T-cells". ASU - Ask A Biologist. 16 February, 2011. https://askabiologist.asu.edu/t-cell
- Hoehn K, et al. The diversity and molecular evolution of B-cell receptors during infection. Mol Biol Evol 33: 1147–1157 (2016). doi: 10.1093/molbev/msw015
- Lythe G, et al. How many TCR clonotypes does a body maintain? J Theor Biol 389: 214–24 (2016). doi: 10.1016/j.jtbi.2015.10.016
- https://www.cancer.gov/publications/dictionaries/cancer-drug/def/anti-pd-1-monoclonal-antibody-medi0680
- Mackensen A, et al. Anti-CD19 CAR T cell therapy for refractory systemic lupus erythematosus. Nat Med 28: 2124–2132 (2022). doi: 10.1038/s41591-022-02017-5
- DeMaio A, et al. The role of the adaptive immune system and T cell dysfunction in neurodegenerative diseases. J Neuroinflammation 19: 251 (2022). doi: 10.1186/s12974-022-02605-9
- Mitra S and Tomar PC. Hybridoma technology; advancements, clinical significance, and future aspects. J Genet Eng Biotechnol 19: 159 (2021). doi: 10.1186/s43141-021-00264-6
About the author:

